6OJ2
 
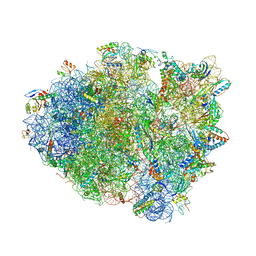 | | Crystal structure of tRNA^ Ala(GGC) bound to the near-cognate 70S A-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-10 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OLA
 
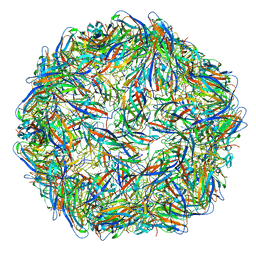 | | Structure of the PCV2d virus-like particle | | 分子名称: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | 著者 | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | 登録日 | 2019-04-16 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
6OLE
 
 | |
6OLF
 
 | |
6OLG
 
 | |
6OLI
 
 | |
6OLJ
 
 | |
6OLZ
 
 | |
6OM0
 
 | |
6OM6
 
 | | Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Mehrani, A, Keiler, K.C, Stagg, S.M, Dunham, C.M. | | 登録日 | 2019-04-18 | | 公開日 | 2021-02-10 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | trans-Translation inhibitors bind to a novel site on the ribosome and clear Neisseria gonorrhoeae in vivo.
Nat Commun, 12, 2021
|
|
6OM7
 
 | |
6OPE
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to near-cognate 70S A site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ORD
 
 | | Crystal structure of tRNA^ Ala(GGC) U32-A38 bound to cognate 70S A site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Nguyen, H.A, Sunita, S, Dunham, C.M. | | 登録日 | 2019-04-30 | | 公開日 | 2020-06-24 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Disruption of evolutionarily correlated tRNA elements impairs accurate decoding.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ORE
 
 | | Release complex 70S | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z. | | 登録日 | 2019-04-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORL
 
 | | RF1 pre-accommodated 70S complex at 24 ms | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-04-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6ORV
 
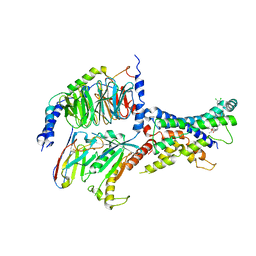 | | Non-peptide agonist (TT-OAD2) bound to the Glucagon-Like peptide-1 (GLP-1) Receptor | | 分子名称: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Belousoff, M.J, Liang, Y.L, Danev, R. | | 登録日 | 2019-05-01 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Activation of the GLP-1 receptor by a non-peptidic agonist.
Nature, 577, 2020
|
|
6OSI
 
 | | Unmodified tRNA(Pro) bound to Thermus thermophilus 70S (near cognate) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hoffer, E.D, Subaramanian, S, Hong, S, Maehigashi, T, Dunham, C.M. | | 登録日 | 2019-05-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (4.14 Å) | | 主引用文献 | Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing.
Elife, 9, 2020
|
|
6OSK
 
 | | RF1 accommodated 70S complex at 60 ms | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-01 | | 公開日 | 2019-06-26 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-26 | | 最終更新日 | 2025-02-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-19 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSY
 
 | |







