8XAB
 
 | |
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
8HN4
 
 | |
8HVK
 
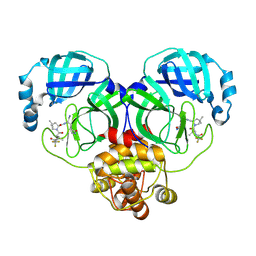 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVL
 
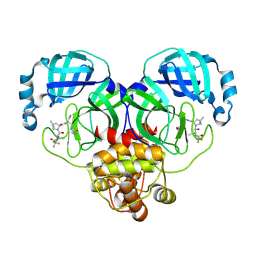 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVM
 
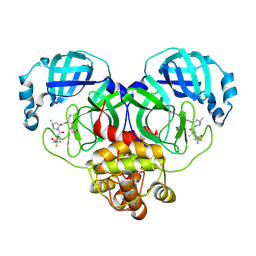 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVN
 
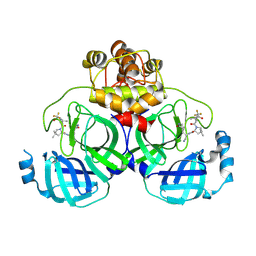 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8HVO
 
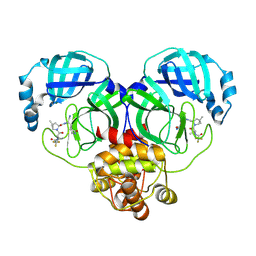 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8I4E
 
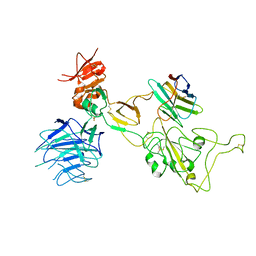 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4F
 
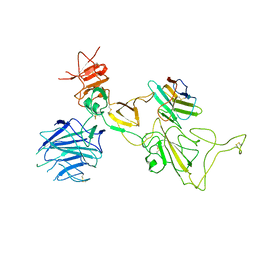 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
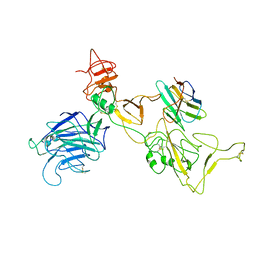 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8JJE
 
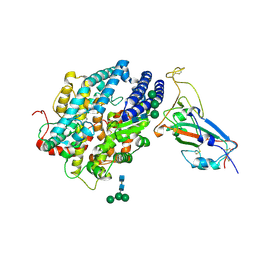 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
8K5G
 
 | | Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD | | Descriptor: | Spike protein S1, UT28K-RD Fab Heavy chain, UT28K-RD Fab Light chain | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
8K5H
 
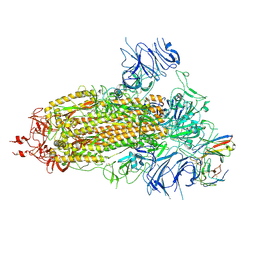 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
8TH1
 
 | |
8IHO
 
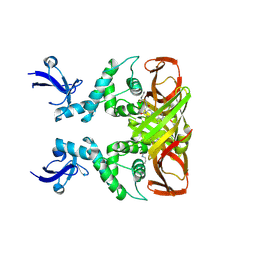 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
8JYK
 
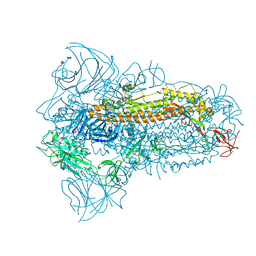 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8JYN
 
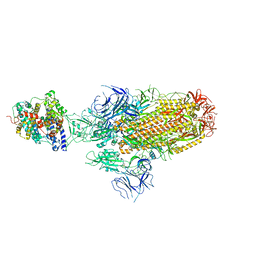 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant.
Nat Commun, 15, 2024
|
|
8FHY
 
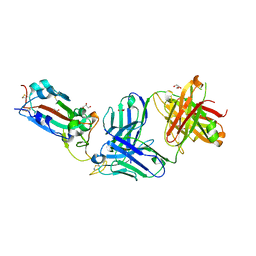 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MALONATE ION, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
8FI9
 
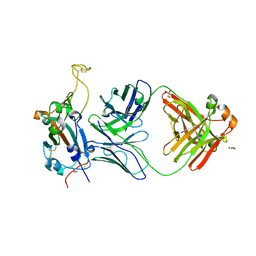 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody WRAIR-5001 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2022-12-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Diverse array of neutralizing antibodies elicited upon Spike Ferritin Nanoparticle vaccination in rhesus macaques.
Nat Commun, 15, 2024
|
|
8HZR
 
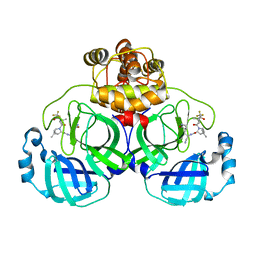 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evaluation of the Inhibition Potency of Nirmatrelvir against Main Protease Mutants of SARS-CoV-2 Variants.
Biochemistry, 62, 2023
|
|
8TM1
 
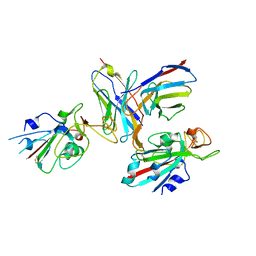 | |
8VCI
 
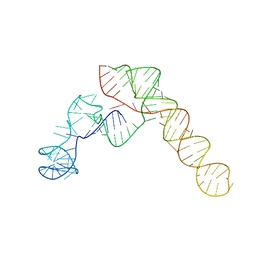 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
8G72
 
 | | SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A) | | Descriptor: | Nanosota-2, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|








