7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | 著者 | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-08-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7K5I
 
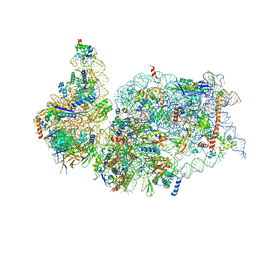 | |
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | 著者 | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-30 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7A29
 
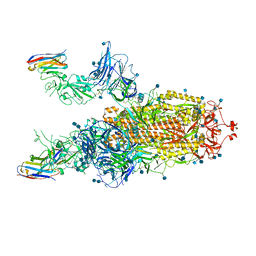 | |
7K8M
 
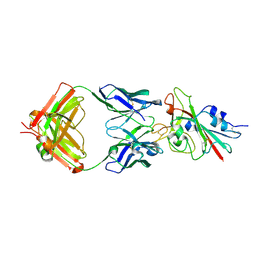 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | 著者 | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8S
 
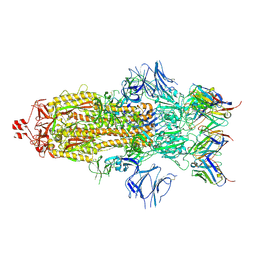 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | 著者 | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8U
 
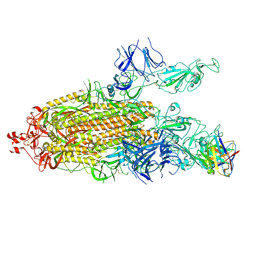 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C104 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C104 Fab Heavy Chain, ... | | 著者 | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
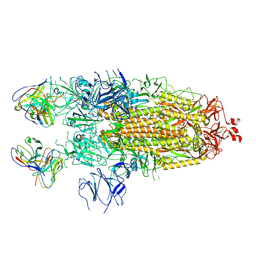 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | 著者 | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
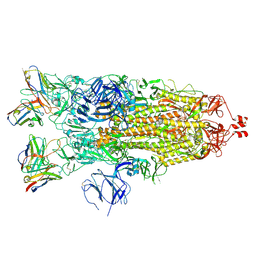 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | 著者 | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8X
 
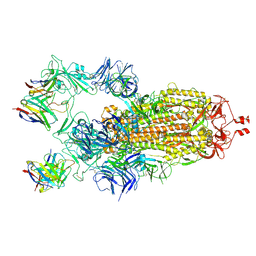 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C121 (State 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C121 Fab Heavy chain, C121 Fab Light chain, ... | | 著者 | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Z
 
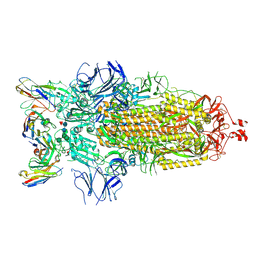 | |
7K90
 
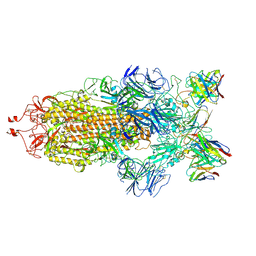 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | 著者 | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K9P
 
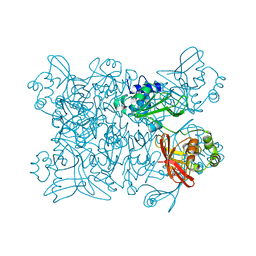 | | Room temperature structure of NSP15 Endoribonuclease from SARS CoV-2 solved using SFX. | | 分子名称: | CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Botha, S, Jernigan, R, Chen, J, Coleman, M.A, Frank, M, Grant, T.D, Hansen, D.T, Ketawala, G, Logeswaran, D, Martin-Garcia, J, Nagaratnam, N, Raj, A.L.L.X, Shelby, M, Yang, J.-H, Yung, M.C, Fromme, P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Structure, 2022
|
|
7KDT
 
 | |
7ACT
 
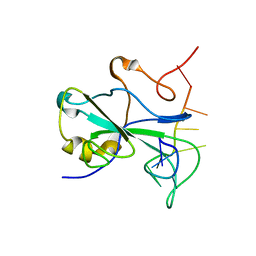 | |
7JJC
 
 | |
7K9Z
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | 著者 | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
7KFI
 
 | | SARS-CoV-2 Main protease immature form - apo structure | | 分子名称: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | 登録日 | 2020-10-14 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | 著者 | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | 登録日 | 2020-10-15 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7A4N
 
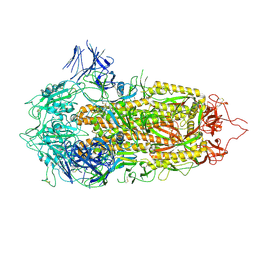 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | 登録日 | 2020-08-20 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
7ACS
 
 | |
7CXM
 
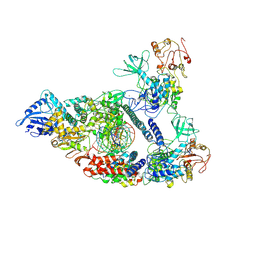 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | 分子名称: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | 登録日 | 2020-09-02 | | 公開日 | 2020-11-04 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7D6H
 
 | |
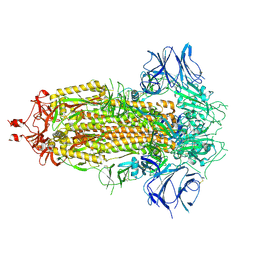
7KDG
 
 | |
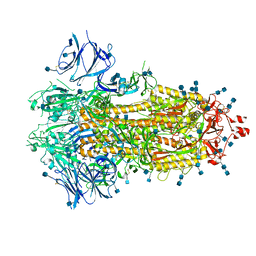
7KDI
 
 | |








