7QEZ
 
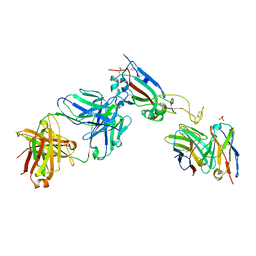 | |
7QF0
 
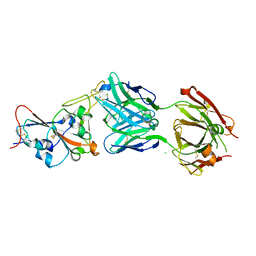 | | Crystal structure of the SARS-CoV-2 RBD in complex with the human antibody CV2.2325 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CV2.2325 heavy chain, ... | | Authors: | Fernandez, I, Pederzoli, R, Rey, F.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2.
J.Exp.Med., 219, 2022
|
|
7QF1
 
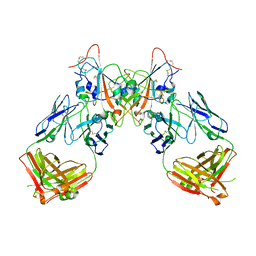 | | Crystal structure of the SARS-CoV-2 RBD in complex with the human antibody CV2.6264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV2.6264 heavy chain, CV2.6264 light chain, ... | | Authors: | Fernandez, I, Pederzoli, R, Rey, F.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2.
J.Exp.Med., 219, 2022
|
|
7WG6
 
 | | Neutral Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z, Wang, X. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7ZDQ
 
 | | Cryo-EM structure of Human ACE2 bound to a high-affinity SARS CoV-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Bate, N, Savva, C.G, Moody, P.C.E, Brown, E.A, Schwabe, W.R, Brindle, N.P.J, Ball, J.K, Sale, J.E. | | Deposit date: | 2022-03-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding.
Plos Pathog., 18, 2022
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7MXP
 
 | | Cryo-EM structure of NTD-directed neutralizing antibody LP5 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)][beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Antibody screening at reduced pH enables preferential selection of potently neutralizing antibodies targeting SARS-CoV-2.
Aiche J, 67, 2021
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7WLY
 
 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
7WLZ
 
 | |
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7WHH
 
 | | Crystal structure of SARS-CoV-2 omicron RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Wang, X.Q, Lan, J. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the SARS-CoV-2 Omicron RBD-ACE2 interaction.
Cell Res., 32, 2022
|
|
7WS0
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS1
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS2
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS3
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS4
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS5
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS6
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 510A5 heavy chain, 510A5 light chain, Spike protein S1 | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS7
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 510A5 heavy chain, 510A5 light chain, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS8
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|
7WS9
 
 | | Structures of Omicron Spike complexes illuminate broad-spectrum neutralizing antibody development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Guo, H, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Omicron spike complexes and implications for neutralizing antibody development.
Cell Rep, 39, 2022
|
|








