[English] 日本語
 Yorodumi
Yorodumi- PDB-1e66: STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1.0E+66 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | |||||||||
 Components Components | ACETYLCHOLINESTERASE | |||||||||
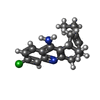
 Keywords Keywords | HYDROLASE / CHOLINESTERASE / HUPRINE X / ALZHEIMER'S DISEASE / CHEMICAL HYBRID | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine catabolic process in synaptic cleft / acetylcholinesterase / choline metabolic process / acetylcholinesterase activity / synaptic cleft / side of membrane / synapse / extracellular space / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | |||||||||
 Authors Authors | Dvir, H. / Harel, M. / Silman, I. / Sussman, J.L. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates. Authors: Dvir, H. / Wong, D.M. / Harel, M. / Barril, X. / Orozco, M. / Luque, F.J. / Munoz-Torrero, D. / Camps, P. / Rosenberry, T.L. / Silman, I. / Sussman, J.L. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1e66.cif.gz 1e66.cif.gz | 136 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1e66.ent.gz pdb1e66.ent.gz | 103.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1e66.json.gz 1e66.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e6/1e66 https://data.pdbj.org/pub/pdb/validation_reports/e6/1e66 ftp://data.pdbj.org/pub/pdb/validation_reports/e6/1e66 ftp://data.pdbj.org/pub/pdb/validation_reports/e6/1e66 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2aceS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 61325.090 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: SYNTHETIC HYBRID, HUPRINE X, BOUND AT THE BOTTOM OF THE ACTIVE SITE GORGE Source: (natural)  Organ: ELECTRIC ORGAN / Variant: G2 FORM / Tissue: ELECTROPLAQUE / References: UniProt: P04058, acetylcholinesterase | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Sugar | | #3: Chemical | ChemComp-HUX / | #4: Water | ChemComp-HOH / | Compound details | HYDROLYZES CHOLINE RELEASED INTO THE SYNAPSE. CATALYTIC ACTIVITY: ACETYLCHOLINE + H(2)O = CHOLINE + ...HYDROLYZES | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.8 Å3/Da / Density % sol: 68 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / pH: 5.6 Details: PROTEIN WAS CRYSTALLISED FROM 35-40% W/V PEG 200 0.3M MES PH 5.6 4 DEG. CELSIUS | |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 6.5 / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X12C / Wavelength: 0.98 / Beamline: X12C / Wavelength: 0.98 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Aug 15, 1999 / Details: MIRRORS |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→29.42 Å / Num. obs: 60094 / % possible obs: 76 % / Redundancy: 11.6 % / Biso Wilson estimate: 9.2 Å2 / Rsym value: 0.061 / Net I/σ(I): 16 |
| Reflection shell | Resolution: 2.1→2.18 Å / Redundancy: 3.52 % / Mean I/σ(I) obs: 2.24 / Rsym value: 0.27 / % possible all: 17.6 |
| Reflection | *PLUS Num. all: 60094 / Num. obs: 45140 / Num. measured all: 482340 / Rmerge(I) obs: 0.06 |
| Reflection shell | *PLUS % possible obs: 20 % / Rmerge(I) obs: 0.27 / Mean I/σ(I) obs: 2.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2ACE Resolution: 2.1→29.42 Å / Rfactor Rfree error: 0.003 / Data cutoff high absF: 2910778.11 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 Details: PROLINE 485 IS OUT OF THE 3FO-2FC DENSITY MAP. THE CONSERVED WATER MOLECULE THAT BELONGS TO THE OXYANION HOLE (HOH 682 IN 2ACE NUMBERING) IS ABSENT IN THIS ENTRY ALTHOUGH A POSITIVE ...Details: PROLINE 485 IS OUT OF THE 3FO-2FC DENSITY MAP. THE CONSERVED WATER MOLECULE THAT BELONGS TO THE OXYANION HOLE (HOH 682 IN 2ACE NUMBERING) IS ABSENT IN THIS ENTRY ALTHOUGH A POSITIVE DIFFERENCE DENSITY HAD BEEN DETECTED IN THAT LOCATION. HERE THIS WATER WAS NOT MODELED BECAUSE OF INSUFICIENT SPACE FOR IT DUE TO MOVEMENT OF SER 200.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 70.6596 Å2 / ksol: 0.345552 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 34.4 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→29.42 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.23 Å / Rfactor Rfree error: 0.016 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.1 Å / Lowest resolution: 40 Å / Rfactor obs: 0.176 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.235 |
 Movie
Movie Controller
Controller



















































 PDBj
PDBj