-Search query
-Search result
Showing all 39 items for (author: xu & py)

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike
EMDB-40985: 
Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state
EMDB-40986: 
Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state
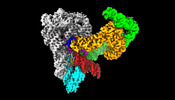
EMDB-40987: 
Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state

PDB-8t2r: 
Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state

PDB-8t2s: 
Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state

PDB-8t2t: 
Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state

EMDB-29910: 
SARS-CoV-2 Spike H655Y variant, One RBD Open
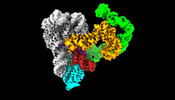
EMDB-26549: 
CryoEM Structure of an Group II Intron Retroelement (apo-complex)
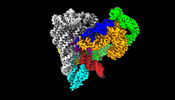
EMDB-26550: 
CryoEM Structure of an Group II Intron Retroelement

PDB-7uim: 
CryoEM Structure of an Group II Intron Retroelement (apo-complex)

PDB-7uin: 
CryoEM Structure of an Group II Intron Retroelement
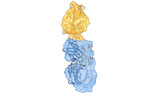
EMDB-33506: 
RBD in complex with Fab14

EMDB-27690: 
Cryo-EM structure of spike binding to Fab of neutralizing antibody (locally refined)

EMDB-31879: 
Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GLP-1R-Gs complex

EMDB-31603: 
Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex

EMDB-31606: 
Cryo-EM structure of the tirzepatide-bound human GIPR-Gs complex

EMDB-31676: 
Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex

EMDB-31836: 
Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GIPR-Gs complex

EMDB-31880: 
Cryo-EM structure of the non-acylated tirzepatide (LY3298176)-bound human GLP-1R-Gs complex

EMDB-31604: 
Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GIPR-Gs complex

EMDB-13075: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450

EMDB-13076: 
STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir

EMDB-13077: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir

PDB-7ouf: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450

PDB-7oug: 
STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir

PDB-7ouh: 
Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir

EMDB-30228: 
Varicella-zoster virus capsid

EMDB-21249: 
Cryo-EM structure of an activated VIP1 receptor-G protein complex

EMDB-10213: 
Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to RBM39 and Indisulam

EMDB-6640: 
cryo-EM map of the full-length human NPC1 at 4.4 angstrom

EMDB-6641: 
cryo-EM map of the full-length human NPC1 at 6.7 angstrom

EMDB-5834: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union

EMDB-5835: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union

EMDB-5836: 
A Unique Human Mycoplasma Protein that Generically Blocks Antigen-Antibody Union

EMDB-1191: 
Conformational changes in the AAA ATPase p97-p47 adaptor complex.

EMDB-1192: 
Conformational changes in the AAA ATPase p97-p47 adaptor complex.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
