[English] 日本語
 Yorodumi
Yorodumi- EMDB-13077: Structure of the STLV intasome:B56 complex bound to the strand-tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13077 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | |||||||||||||||
 Map data Map data | DeepEMenhancer highest target map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | integrase / intasome / HTLV / STLV / integration / strand-transfer inhibitors / INSTI / bictegravir / BIC / drug / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / DNA recombination / symbiont entry into host cell / zinc ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Simian T-lymphotropic virus 1 / Simian T-lymphotropic virus 1 /  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Barski MS / Ballandras-Colas A | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures. Authors: Michal S Barski / Teresa Vanzo / Xue Zhi Zhao / Steven J Smith / Allison Ballandras-Colas / Nora B Cronin / Valerie E Pye / Stephen H Hughes / Terrence R Burke / Peter Cherepanov / Goedele N Maertens /     Abstract: Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this ...Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this deltaretrovirus. Here, we screened a library of integrase strand transfer inhibitor (INSTI) candidates built around several chemical scaffolds to determine their effectiveness in limiting HTLV-1 infection. Naphthyridines with substituents in position 6 emerged as the most potent compounds against HTLV-1, with XZ450 having highest efficacy in vitro. Using single-particle cryo-electron microscopy we visualised XZ450 as well as the clinical HIV-1 INSTIs raltegravir and bictegravir bound to the active site of the deltaretroviral intasome. The structures reveal subtle differences in the coordination environment of the Mg ion pair involved in the interaction with the INSTIs. Our results elucidate the binding of INSTIs to the HTLV-1 intasome and support their use for pre-exposure prophylaxis and possibly future treatment of HTLV-1 infection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13077.map.gz emd_13077.map.gz | 92.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13077-v30.xml emd-13077-v30.xml emd-13077.xml emd-13077.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13077_fsc.xml emd_13077_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13077.png emd_13077.png | 110.8 KB | ||
| Filedesc metadata |  emd-13077.cif.gz emd-13077.cif.gz | 8.3 KB | ||
| Others |  emd_13077_additional_1.map.gz emd_13077_additional_1.map.gz emd_13077_half_map_1.map.gz emd_13077_half_map_1.map.gz emd_13077_half_map_2.map.gz emd_13077_half_map_2.map.gz | 6.7 MB 82.7 MB 82.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13077 http://ftp.pdbj.org/pub/emdb/structures/EMD-13077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13077 | HTTPS FTP |
-Related structure data
| Related structure data |  7ouhMC  7oufC  7ougC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13077.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13077.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMenhancer highest target map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: denmod map - model was refined against this map
| File | emd_13077_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | denmod map - model was refined against this map | ||||||||||||
| Projections & Slices |
| ||||||||||||
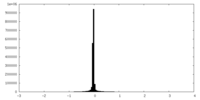
| Density Histograms |
-Half map: half map 1
| File | emd_13077_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_13077_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of STLV-1 MarB43 integrase with nascent viral DNA, the hu...
+Supramolecule #1: Complex of STLV-1 MarB43 integrase with nascent viral DNA, the hu...
+Supramolecule #2: Integrase
+Supramolecule #3: PC4 and SFRS1-interacting protein,Serine/threonine-protein phosph...
+Supramolecule #4: DNA
+Macromolecule #1: Integrase
+Macromolecule #2: PC4 and SFRS1-interacting protein,Serine/threonine-protein phosph...
+Macromolecule #3: DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*...
+Macromolecule #4: DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*...
+Macromolecule #5: ZINC ION
+Macromolecule #6: MAGNESIUM ION
+Macromolecule #7: Bictegravir
+Macromolecule #8: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. Details: Glow-discharged for 4 min at 45 mA on an Emitech K100X instrument (Electron Microscopy Sciences) and covered with a layer of graphene oxide (Sigma-Aldrich, catalogue #763705) immediately before being used. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Complex was isolated by size exclusion chromatography |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID:  6z2y Chain - Source name: PDB / Chain - Initial model type: experimental model Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix. ...Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix.real_space_refine using C2 NCS and secondary structure and metal ion coordination restraints. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-7ouh: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)