[English] 日本語
 Yorodumi
Yorodumi- PDB-7ouh: Structure of the STLV intasome:B56 complex bound to the strand-tr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7ouh | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | VIRAL PROTEIN / integrase / intasome / HTLV / STLV / integration / strand-transfer inhibitors / INSTI / bictegravir / BIC / drug | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA stem-loop binding / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / DNA recombination / symbiont entry into host cell / zinc ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1 Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Barski, M.S. / Ballandras-Colas, A. / Cronin, N.B. / Pye, V.E. / Cherepanov, P. / Maertens, G.N. | |||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  United States, 4items United States, 4items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures. Authors: Michal S Barski / Teresa Vanzo / Xue Zhi Zhao / Steven J Smith / Allison Ballandras-Colas / Nora B Cronin / Valerie E Pye / Stephen H Hughes / Terrence R Burke / Peter Cherepanov / Goedele N Maertens /     Abstract: Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this ...Between 10 and 20 million people worldwide are infected with the human T-cell lymphotropic virus type 1 (HTLV-1). Despite causing life-threatening pathologies there is no therapeutic regimen for this deltaretrovirus. Here, we screened a library of integrase strand transfer inhibitor (INSTI) candidates built around several chemical scaffolds to determine their effectiveness in limiting HTLV-1 infection. Naphthyridines with substituents in position 6 emerged as the most potent compounds against HTLV-1, with XZ450 having highest efficacy in vitro. Using single-particle cryo-electron microscopy we visualised XZ450 as well as the clinical HIV-1 INSTIs raltegravir and bictegravir bound to the active site of the deltaretroviral intasome. The structures reveal subtle differences in the coordination environment of the Mg ion pair involved in the interaction with the INSTIs. Our results elucidate the binding of INSTIs to the HTLV-1 intasome and support their use for pre-exposure prophylaxis and possibly future treatment of HTLV-1 infection. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7ouh.cif.gz 7ouh.cif.gz | 362.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7ouh.ent.gz pdb7ouh.ent.gz | 276.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7ouh.json.gz 7ouh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ou/7ouh https://data.pdbj.org/pub/pdb/validation_reports/ou/7ouh ftp://data.pdbj.org/pub/pdb/validation_reports/ou/7ouh ftp://data.pdbj.org/pub/pdb/validation_reports/ou/7ouh | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13077MC  7oufC  7ougC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 2 types, 6 molecules DEABFC
| #1: Protein | Mass: 33943.539 Da / Num. of mol.: 4 / Mutation: A219E Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Simian T-lymphotropic virus 1 / Gene: pol / Production host: Simian T-lymphotropic virus 1 / Gene: pol / Production host:  #2: Protein | Mass: 80394.680 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no ...Details: fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no space for these details below),fusion construct containing human LEDGF (residues 1-324 thus without the IBD domain) (gene PSIP1; O75475)) and human B56gammma (residues 11-380) regulatory subunit of PP2A (gene PPP2R5C) (no space for these details below) Source: (gene. exp.)  Homo sapiens (human) / Gene: PSIP1, DFS70, LEDGF, PSIP2, PPP2R5C, KIAA0044 / Production host: Homo sapiens (human) / Gene: PSIP1, DFS70, LEDGF, PSIP2, PPP2R5C, KIAA0044 / Production host:  |
|---|
-DNA chain , 2 types, 4 molecules IKJL
| #3: DNA chain | Mass: 9221.909 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1#4: DNA chain | Mass: 8593.560 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Simian T-lymphotropic virus 1 Simian T-lymphotropic virus 1 |
|---|
-Non-polymers , 4 types, 16 molecules 

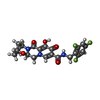




| #5: Chemical | ChemComp-ZN / #6: Chemical | ChemComp-MG / #7: Chemical | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.3311 MDa / Experimental value: NO | |||||||||||||||||||||||||||||||||||
| Source (natural) |
| |||||||||||||||||||||||||||||||||||
| Source (recombinant) |
| |||||||||||||||||||||||||||||||||||
| Buffer solution | pH: 6 | |||||||||||||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||||||||||||
| Specimen | Conc.: 0.7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: Complex was isolated by size exclusion chromatography | |||||||||||||||||||||||||||||||||||
| Specimen support | Details: Glow-discharged for 4 min at 45 mA on an Emitech K100X instrument (Electron Microscopy Sciences) and covered with a layer of graphene oxide (Sigma-Aldrich, catalogue #763705) immediately before being used. Grid material: GOLD / Grid type: UltrAuFoil R1.2/1.3 | |||||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 95 % / Chamber temperature: 295.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software | Name: PHENIX / Version: dev_4155: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1714199 | ||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C2 (2 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 39731 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6Z2Y 6z2y Accession code: 6Z2Y Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix. ...Details: 6Z2Y was fitted into the cryoEM map using Chimera. The model was adjusted to fit the map; metal ions and drug docked into the map manually using Coot. The final model was subjected to Phenix.real_space_refine using C2 NCS and secondary structure and metal ion coordination restraints. Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj









































