+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Antibody N3-1 bound to SARS-CoV-2 spike | |||||||||
 Map data Map data | sharpen map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 spike / neutralizing antibody / RBD-directed antibody / quaternary epitope / VIRAL PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Hsieh C-L / McLellan JS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: SARS-COV-2 Omicron variants conformationally escape a rare quaternary antibody binding mode. Authors: Jule Goike / Ching-Lin Hsieh / Andrew P Horton / Elizabeth C Gardner / Ling Zhou / Foteini Bartzoka / Nianshuang Wang / Kamyab Javanmardi / Andrew Herbert / Shawn Abbassi / Xuping Xie / ...Authors: Jule Goike / Ching-Lin Hsieh / Andrew P Horton / Elizabeth C Gardner / Ling Zhou / Foteini Bartzoka / Nianshuang Wang / Kamyab Javanmardi / Andrew Herbert / Shawn Abbassi / Xuping Xie / Hongjie Xia / Pei-Yong Shi / Rebecca Renberg / Thomas H Segall-Shapiro / Cynthia I Terrace / Wesley Wu / Raghav Shroff / Michelle Byrom / Andrew D Ellington / Edward M Marcotte / James M Musser / Suresh V Kuchipudi / Vivek Kapur / George Georgiou / Scott C Weaver / John M Dye / Daniel R Boutz / Jason S McLellan / Jimmy D Gollihar /  Abstract: The ongoing evolution of SARS-CoV-2 into more easily transmissible and infectious variants has provided unprecedented insight into mutations enabling immune escape. Understanding how these mutations ...The ongoing evolution of SARS-CoV-2 into more easily transmissible and infectious variants has provided unprecedented insight into mutations enabling immune escape. Understanding how these mutations affect the dynamics of antibody-antigen interactions is crucial to the development of broadly protective antibodies and vaccines. Here we report the characterization of a potent neutralizing antibody (N3-1) identified from a COVID-19 patient during the first disease wave. Cryogenic electron microscopy revealed a quaternary binding mode that enables direct interactions with all three receptor-binding domains of the spike protein trimer, resulting in extraordinary avidity and potent neutralization of all major variants of concern until the emergence of Omicron. Structure-based rational design of N3-1 mutants improved binding to all Omicron variants but only partially restored neutralization of the conformationally distinct Omicron BA.1. This study provides new insights into immune evasion through changes in spike protein dynamics and highlights considerations for future conformationally biased multivalent vaccine designs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41399.map.gz emd_41399.map.gz | 290.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41399-v30.xml emd-41399-v30.xml emd-41399.xml emd-41399.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41399.png emd_41399.png | 99 KB | ||
| Filedesc metadata |  emd-41399.cif.gz emd-41399.cif.gz | 6.4 KB | ||
| Others |  emd_41399_additional_1.map.gz emd_41399_additional_1.map.gz emd_41399_additional_2.map.gz emd_41399_additional_2.map.gz emd_41399_additional_3.map.gz emd_41399_additional_3.map.gz emd_41399_half_map_1.map.gz emd_41399_half_map_1.map.gz emd_41399_half_map_2.map.gz emd_41399_half_map_2.map.gz | 154.1 MB 270.9 MB 251.5 MB 285.8 MB 285.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41399 http://ftp.pdbj.org/pub/emdb/structures/EMD-41399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41399 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41399.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41399.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpen map | ||||||||||||||||||||||||||||||||||||
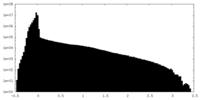
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
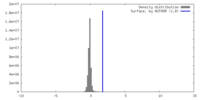
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: map
| File | emd_41399_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DeepEMhancer map
| File | emd_41399_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Composite map
| File | emd_41399_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_41399_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_41399_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of N3-1 bound to SARS-CoV-2 spike
| Entire | Name: Complex of N3-1 bound to SARS-CoV-2 spike |
|---|---|
| Components |
|
-Supramolecule #1: Complex of N3-1 bound to SARS-CoV-2 spike
| Supramolecule | Name: Complex of N3-1 bound to SARS-CoV-2 spike / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: SARS-CoV-2 spike
| Supramolecule | Name: SARS-CoV-2 spike / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: N3-1
| Supramolecule | Name: N3-1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SARS-CoV-2 spike
| Macromolecule | Name: SARS-CoV-2 spike / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLT T RTQLPPAY TN SFTRGVY YPD KVFRSS VLHS TQDLF LPFFS NVTW FHAIHV SGT NGTKRFD NP VLPFNDGV Y FASTEKSNI IRGWIFGTTL DSKTQSLLI V NNATNVVI KV CEFQFCN DPF LGVYYH KNNK SWMES ...String: MFVFLVLLPL VSSQCVNLT T RTQLPPAY TN SFTRGVY YPD KVFRSS VLHS TQDLF LPFFS NVTW FHAIHV SGT NGTKRFD NP VLPFNDGV Y FASTEKSNI IRGWIFGTTL DSKTQSLLI V NNATNVVI KV CEFQFCN DPF LGVYYH KNNK SWMES EFRVY SSAN NCTFEY VSQ PFLMDLE GK QGNFKNLR E FVFKNIDGY FKIYSKHTPI NLVRDLPQG F SALEPLVD LP IGINITR FQT LLALHR SYLT PGDSS SGWTA GAAA YYVGYL QPR TFLLKYN EN GTITDAVD C ALDPLSETK CTLKSFTVEK GIYQTSNFR V QPTESIVR FP NITNLCP FGE VFNATR FASV YAWNR KRISN CVAD YSVLYN SAS FSTFKCY GV SPTKLNDL C FTNVYADSF VIRGDEVRQI APGQTGKIA D YNYKLPDD FT GCVIAWN SNN LDSKVG GNYN YLYRL FRKSN LKPF ERDIST EIY QAGSTPC NG VEGFNCYF P LQSYGFQPT NGVGYQPYRV VVLSFELLH A PATVCGPK KS TNLVKNK CVN FNFNGL TGTG VLTES NKKFL PFQQ FGRDIA DTT DAVRDPQ TL EILDITPC S FGGVSVITP GTNTSNQVAV LYQDVNCTE V PVAIHADQ LT PTWRVYS TGS NVFQTR AGCL IGAEH VNNSY ECDI PIGAGI CAS YQTQTNS PG SASSVASQ S IIAYTMSLG AENSVAYSNN SIAIPTNFT I SVTTEILP VS MTKTSVD CTM YICGDS TECS NLLLQ YGSFC TQLN RALTGI AVE QDKNTQE VF AQVKQIYK T PPIKDFGGF NFSQILPDPS KPSKRSPIE D LLFNKVTL AD AGFIKQY GDC LGDIAA RDLI CAQKF NGLTV LPPL LTDEMI AQY TSALLAG TI TSGWTFGA G PALQIPFPM QMAYRFNGIG VTQNVLYEN Q KLIANQFN SA IGKIQDS LSS TPSALG KLQD VVNQN AQALN TLVK QLSSNF GAI SSVLNDI LS RLDPPEAE V QIDRLITGR LQSLQTYVTQ QLIRAAEIR A SANLAATK MS ECVLGQS KRV DFCGKG YHLM SFPQS APHGV VFLH VTYVPA QEK NFTTAPA IC HDGKAHFP R EGVFVSNGT HWFVTQRNFY EPQIITTDN T FVSGNCDV VI GIVNNTV YDP LQPELD SFKE ELDKY FKNHT SPDV DLGDIS GIN ASVVNIQ KE IDRLNEVA K NLNESLIDL QELGKYEQGS GYIPEAPRD G QAYVRKDG EW VLLSTFL GRS LEVLFQ GPGH HHHHH HHSAW SHPQ FEKGGG SGG GGSGGSA WS HPQFEK |
-Macromolecule #2: N3-1 Fab heavy chain
| Macromolecule | Name: N3-1 Fab heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQWGPG LVNPSETLSL TCSVSGGSFA TENYYWSWIR QHPGEGLEWI GNIYFSGNTY YNPSLNNRFT ISFDTSKNHL SLKLPSVTAA DTAVYYCARG TIYFDRSGYR RVDPFHIWGQ GTMVIVSS |
-Macromolecule #3: N3-1 Fab light chain
| Macromolecule | Name: N3-1 Fab light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPST LSASVGDRVT ITCRASQSIS SWLAWYQQKP GKAPKLLIYD ASSLESGVPS RFSGSGSGTE FTLTISSLQP DDFATYYCQQ YNSYSPWTFG QGTKVEIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 2 mM Tris pH 8.0, 200 mM NaCl, 0.02% NaN3 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
| Details | Collected on UltrAuFoil 1.2-1.3 with 0.2 mg/mL HexaPro and 5x fold molar excess FabN3-1 spiked in 30 minutes before freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 266585 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)