-Search query
-Search result
Showing 1 - 50 of 119 items for (author: wang & zw)

EMDB-64784: 
Structure of TolC and YbjP closed state complex
Method: single particle / : Ge XF, Gu ZW, Wang JW

EMDB-64785: 
Structure of TolC, YbjP, and AcrABZ complex
Method: single particle / : Ge XF, Gu ZW, Wang JW

EMDB-64787: 
Structure of TolC, YbjP, and AcrA complex
Method: single particle / : Ge XF, Gu ZW, Wang JW

EMDB-62490: 
Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II
Method: single particle / : Li ZW, Ye Y, Yang GF

EMDB-62491: 
Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II in UQ1-bound state
Method: single particle / : Li ZW, Ye Y, Yang GF

EMDB-62495: 
Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II in pydiflumetofen-bound state
Method: single particle / : Li ZW, Ye Y, Yang GF

EMDB-63115: 
Cryo-EM structure of Saccharomyces cerevisiae Mitochondrial Respiratory Complex II in Y19315-bound state
Method: single particle / : Li ZW, Ye Y, Yang GF

EMDB-65574: 
In situ structure of mitochondrial ATP synthase in whole C. reinhardtii cells
Method: subtomogram averaging / : Luo ZW, Wang QH, Ma JP

EMDB-65576: 
In situ structure of S. pombe 80S ribosome
Method: subtomogram averaging / : Luo ZW, Wang QH, Ma JP

EMDB-65645: 
In situ structure of S. pombe Fatty Acid Synthase
Method: subtomogram averaging / : Luo ZW, Wang QH, Ma JP

EMDB-61620: 
Cryo-EM structure of human SLFN14
Method: single particle / : Luo M, Jia XD, Wang ZW, Yang JY, Zhang QF, Gao S

EMDB-47091: 
Taeniopygia guttata R2 retrotransposon (R2Tg) initiating target-primed reverse transcription
Method: single particle / : Wilkinson ME, Edmonds KHK, Zhang F

PDB-9dou: 
Taeniopygia guttata R2 retrotransposon (R2Tg) initiating target-primed reverse transcription
Method: single particle / : Wilkinson ME, Edmonds KHK, Zhang F

EMDB-44627: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1533 (local refinement of NTD and C1533)
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO

EMDB-44628: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO

EMDB-44629: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO

PDB-9bj2: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1533 (local refinement of NTD and C1533)
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO

PDB-9bj3: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO

PDB-9bj4: 
Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952
Method: single particle / : Rubio AA, Abernathy ME, Barnes CO
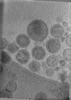
EMDB-47705: 
Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J
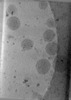
EMDB-47741: 
Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47743: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47745: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J
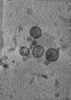
EMDB-47858: 
Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-61073: 
Cap region of monocin
Method: single particle / : Wang JW, Gu ZW

EMDB-61074: 
Tip region of monocin
Method: single particle / : Wang JW, Gu ZW

EMDB-61075: 
Side fiber of monocin
Method: single particle / : Wang JW, Gu ZW

EMDB-61387: 
Structure of chanoclavine synthase from Claviceps fusiformis
Method: single particle / : Liu ZW, Wang T, Li X, Shen PP, Huang JW, Chen CC, Guo RT

EMDB-61388: 
Structure of chanoclavine synthase from Claviceps fusiformis in complex with prechanoclavine
Method: single particle / : Liu ZW, Wang T, Li X, Shen PP, Huang JW, Chen CC, Guo RT

EMDB-39291: 
Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in pyraclostrobin-bound state
Method: single particle / : Ye Y, Li ZW, Yang GF

EMDB-39323: 
Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in YF23694-bound state
Method: single particle / : Ye Y, Li ZW, Yang GF

EMDB-60256: 
Cryo-EM structure of Saccharomyces cerevisiae bc1 complex in Metyltetraprole-bound state
Method: single particle / : Ye Y, Li ZW, Yang GF

EMDB-60317: 
Cryo-EM structure of pyraclostrobin-bound porcine bc1 complex
Method: single particle / : Wang YX, Sun JY, Li ZW, Cui GR, Yang GF

EMDB-60320: 
Cryo-EM structure of Metyltetraprole-bound porcine bc1 complex
Method: single particle / : Wang YX, Sun JY, Cui GR, Yang GF

EMDB-60323: 
Cryo-EM structure of YF23694-bound porcine bc1 complex
Method: single particle / : Wang YX, Sun JY, Cui GR, Yang GF

EMDB-37641: 
multidrug transporter EfpA from Mycobacterium tuberculosis bound with lipids
Method: single particle / : Wang S, Liao M

EMDB-42204: 
Multidrug efflux pump MtEfpA bound with inhibitor BRD8000.3
Method: single particle / : Wang S, Liao M

EMDB-42205: 
Multidrug efflux pump EfpA from mycobacterium smegmatis
Method: single particle / : Wang S, Liao M

EMDB-38540: 
Prohead portal of bacteriophage lambda
Method: single particle / : Wang JW, Gu ZW

EMDB-38541: 
Prohead portal vertex of bacteriophage lambda
Method: single particle / : Wang JW, Gu ZW

EMDB-38542: 
Mature virion portal of bacteriophage lambda
Method: single particle / : Wang JW, Gu ZW

EMDB-38556: 
Mature virion portal of phage lambda with DNA
Method: single particle / : Wang JW, Gu ZW

EMDB-38572: 
Mature virion portal vertex of bacteriophage lambda
Method: single particle / : Wang JW, Gu ZW

EMDB-34200: 
Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus
Method: single particle / : Lu K, Luo B, Tao X, Li H, Xie Y, Zhao Z, Xia W, Su Z, Mao Z

EMDB-35112: 
Cryo-EM map of SepF
Method: single particle / : Zhang HW, Zhang C, Chang ZW, Yang J

EMDB-34948: 
Cryo-EM structure of the apo-GPR132-Gi
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-34950: 
Activation mechanism of GPR132 by NPGLY
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-34951: 
Activation mechanism of GPR132 by 9(S)-HODE
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-35044: 
Activation mechanism of GPR132 by compound NOX-6-7
Method: single particle / : Wang JL, Ding JH, Sun JP, Yu X

EMDB-33942: 
Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2
Method: single particle / : Hsu STD, Chang NE, Weng ZW, Yang TJ, Draczkowski P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
