+Search query
-Structure paper
| Title | Mechanism-guided engineering of a minimal biological particle for genome editing. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 122, Issue 1, Page e2413519121, Year 2025 |
| Publish date | Jan 7, 2025 |
 Authors Authors | Wayne Ngo / Julia Peukes / Alisha Baldwin / Zhiwei Wayne Xue / Sidney Hwang / Robert R Stickels / Zhi Lin / Ansuman T Satpathy / James A Wells / Randy Schekman / Eva Nogales / Jennifer A Doudna /  |
| PubMed Abstract | The widespread application of genome editing to treat and cure disease requires the delivery of genome editors into the nucleus of target cells. Enveloped delivery vehicles (EDVs) are engineered ...The widespread application of genome editing to treat and cure disease requires the delivery of genome editors into the nucleus of target cells. Enveloped delivery vehicles (EDVs) are engineered virally derived particles capable of packaging and delivering CRISPR-Cas9 ribonucleoproteins (RNPs). However, the presence of lentiviral genome encapsulation and replication proteins in EDVs has obscured the underlying delivery mechanism and precluded particle optimization. Here, we show that Cas9 RNP nuclear delivery is independent of the native lentiviral capsid structure. Instead, EDV-mediated genome editing activity corresponds directly to the number of nuclear localization sequences on the Cas9 enzyme. EDV structural analysis using cryo-electron tomography and small molecule inhibitors guided the removal of ~80% of viral residues, creating a minimal EDV (miniEDV) that retains full RNP delivery capability. MiniEDVs are 25% smaller yet package equivalent amounts of Cas9 RNPs relative to the original EDVs and demonstrated increased editing in cell lines and therapeutically relevant primary human T cells. These results show that virally derived particles can be streamlined to create efficacious genome editing delivery vehicles with simpler production and manufacturing. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:39793042 / PubMed:39793042 /  PubMed Central PubMed Central |
| Methods | EM (tomography) / EM (subtomogram averaging) |
| Resolution | 9.5 - 10.5 Å |
| Structure data | 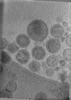 EMDB-47705: Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs) 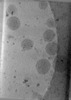 EMDB-47741: Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9  EMDB-47743: Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9  EMDB-47745: Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9 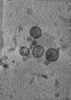 EMDB-47858: Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV) |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 lentivirus derived envelope delivery vehicle
lentivirus derived envelope delivery vehicle