+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
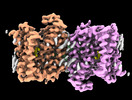
| Title | Multidrug efflux pump MtEfpA bound with inhibitor BRD8000.3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | multidrug efflux pump / EfpA / mycobacterium / BRD8000.3 / TRANSPORT PROTEIN | |||||||||
| Function / homology | Major facilitator superfamily / Major Facilitator Superfamily / Major facilitator superfamily domain / Major facilitator superfamily (MFS) profile. / transmembrane transporter activity / MFS transporter superfamily / plasma membrane / MFS-type transporter EfpA Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
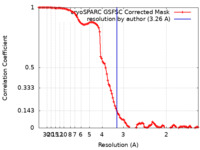
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Wang S / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structures of the Mycobacterium tuberculosis efflux pump EfpA reveal the mechanisms of transport and inhibition. Authors: Shuhui Wang / Kun Wang / Kangkang Song / Zon Weng Lai / Pengfei Li / Dongying Li / Yajie Sun / Ye Mei / Chen Xu / Maofu Liao /   Abstract: As the first identified multidrug efflux pump in Mycobacterium tuberculosis (Mtb), EfpA is an essential protein and promising drug target. However, the functional and inhibitory mechanisms of EfpA ...As the first identified multidrug efflux pump in Mycobacterium tuberculosis (Mtb), EfpA is an essential protein and promising drug target. However, the functional and inhibitory mechanisms of EfpA are poorly understood. Here we report cryo-EM structures of EfpA in outward-open conformation, either bound to three endogenous lipids or the inhibitor BRD-8000.3. Three lipids inside EfpA span from the inner leaflet to the outer leaflet of the membrane. BRD-8000.3 occupies one lipid site at the level of inner membrane leaflet, competitively inhibiting lipid binding. EfpA resembles the related lysophospholipid transporter MFSD2A in both overall structure and lipid binding sites and may function as a lipid flippase. Combining AlphaFold-predicted EfpA structure, which is inward-open, we propose a complete conformational transition cycle for EfpA. Together, our results provide a structural and mechanistic foundation to comprehend EfpA function and develop EfpA-targeting anti-TB drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42204.map.gz emd_42204.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42204-v30.xml emd-42204-v30.xml emd-42204.xml emd-42204.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42204_fsc.xml emd_42204_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42204.png emd_42204.png | 114.3 KB | ||
| Filedesc metadata |  emd-42204.cif.gz emd-42204.cif.gz | 5.6 KB | ||
| Others |  emd_42204_half_map_1.map.gz emd_42204_half_map_1.map.gz emd_42204_half_map_2.map.gz emd_42204_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42204 http://ftp.pdbj.org/pub/emdb/structures/EMD-42204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42204 | HTTPS FTP |
-Related structure data
| Related structure data |  8ufdMC  8ufeC  8wm5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42204.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42204.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.788 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42204_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42204_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Multidrug efflux pump MtEfpA with lipids and inhibitor...
| Entire | Name: Complex of Multidrug efflux pump MtEfpA with lipids and inhibitor BRD8000.3 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Multidrug efflux pump MtEfpA with lipids and inhibitor...
| Supramolecule | Name: Complex of Multidrug efflux pump MtEfpA with lipids and inhibitor BRD8000.3 type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Integral membrane efflux protein EFPA
| Macromolecule | Name: Integral membrane efflux protein EFPA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 55.620098 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTALNDTERA VRNWTAGRPH RPAPMRPPRS EETASERPSR YYPTWLPSRS FIAAVIAIGG MQLLATMDST VAIVALPKIQ NELSLSDAG RSWVITAYVL TFGGLMLLGG RLGDTIGRKR TFIVGVALFT ISSVLCAVAW DEATLVIARL SQGVGSAIAS P TGLALVAT ...String: MTALNDTERA VRNWTAGRPH RPAPMRPPRS EETASERPSR YYPTWLPSRS FIAAVIAIGG MQLLATMDST VAIVALPKIQ NELSLSDAG RSWVITAYVL TFGGLMLLGG RLGDTIGRKR TFIVGVALFT ISSVLCAVAW DEATLVIARL SQGVGSAIAS P TGLALVAT TFPKGPARNA ATAVFAAMTA IGSVMGLVVG GALTEVSWRW AFLVNVPIGL VMIYLARTAL RETNKERMKL DA TGAILAT LACTAAVFAF SIGPEKGWMS GITIGSGLVA LAAAVAFVIV ERTAENPVVP FHLFRDRNRL VTFSAILLAG GVM FSLTVC IGLYVQDILG YSALRAGVGF IPFVIAMGIG LGVSSQLVSR FSPRVLTIGG GYLLFGAMLY GSFFMHRGVP YFPN LVMPI VVGGIGIGMA VVPLTLSAIA GVGFDQIGPV SAIALMLQSL GGPLVLAVIQ AVITSRTLYL GGTTGPVKFM NDVQL AALD HAYTYGLLWV AGAAIIVGGM ALFIGYTPQQ VAHAQEVKEA IDAGEL UniProtKB: MFS-type transporter EfpA |
-Macromolecule #2: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 2 / Number of copies: 4 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #3: (1S,3S)-N-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3...
| Macromolecule | Name: (1S,3S)-N-[6-bromo-5-(pyrimidin-2-yl)pyridin-2-yl]-2,2-dimethyl-3-(2-methylpropyl)cyclopropane-1-carboxamide type: ligand / ID: 3 / Number of copies: 2 / Formula: WJI |
|---|---|
| Molecular weight | Theoretical: 403.316 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)