+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
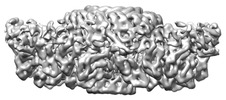
| Title | Prohead portal vertex of bacteriophage lambda | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | caudovirales / siphoviridae / portal vertex / portal / capsid / connector/neck / tail / delivery device / B-DNA / phage lambda / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral portal complex / symbiont genome ejection through host cell envelope, long flexible tail mechanism / T=7 icosahedral viral capsid / virion assembly / viral capsid / host cell cytoplasm / structural molecule activity / DNA binding Similarity search - Function | |||||||||
| Biological species |  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||
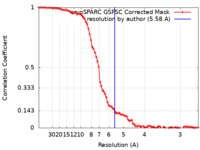
| Method | single particle reconstruction / cryo EM / Resolution: 5.58 Å | |||||||||
 Authors Authors | Wang JW / Gu ZW | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2024 Journal: J Virol / Year: 2024Title: Structural morphing in the viral portal vertex of bacteriophage lambda. Authors: Zhiwei Gu / Kexun Wu / Jiawei Wang /  Abstract: The portal protein of tailed bacteriophage plays essential roles in various aspects of capsid assembly, motor assembly, genome packaging, connector formation, and infection processes. After DNA ...The portal protein of tailed bacteriophage plays essential roles in various aspects of capsid assembly, motor assembly, genome packaging, connector formation, and infection processes. After DNA packaging is complete, additional proteins are assembled onto the portal to form the connector complex, which is crucial as it bridges the mature head and tail. In this study, we report high-resolution cryo-electron microscopy (cryo-EM) structures of the portal vertex from bacteriophage lambda in both its prohead and mature virion states. Comparison of these structures shows that during head maturation, in addition to capsid expansion, the portal protein undergoes conformational changes to establish interactions with the connector proteins. Additionally, the independently assembled tail undergoes morphological alterations at its proximal end, facilitating its connection to the head-tail joining protein and resulting in the formation of a stable portal-connector-tail complex. The B-DNA molecule spirally glides through the tube, interacting with the nozzle blade region of the middle-ring connector protein. These insights elucidate a mechanism for portal maturation and DNA translocation within the phage lambda system. IMPORTANCE: The tailed bacteriophages possess a distinct portal vertex that consists of a ring of 12 portal proteins associated with a 5-fold capsid shell. This portal protein is crucial in multiple ...IMPORTANCE: The tailed bacteriophages possess a distinct portal vertex that consists of a ring of 12 portal proteins associated with a 5-fold capsid shell. This portal protein is crucial in multiple stages of virus assembly and infection. Our research focused on examining the structures of the portal vertex in both its preliminary prohead state and the fully mature virion state of bacteriophage lambda. By analyzing these structures, we were able to understand how the portal protein undergoes conformational changes during maturation, the mechanism by which it prevents DNA from escaping, and the process of DNA spirally gliding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38541.map.gz emd_38541.map.gz | 31 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38541-v30.xml emd-38541-v30.xml emd-38541.xml emd-38541.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38541_fsc.xml emd_38541_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_38541.png emd_38541.png | 67.3 KB | ||
| Filedesc metadata |  emd-38541.cif.gz emd-38541.cif.gz | 5.5 KB | ||
| Others |  emd_38541_half_map_1.map.gz emd_38541_half_map_1.map.gz emd_38541_half_map_2.map.gz emd_38541_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38541 http://ftp.pdbj.org/pub/emdb/structures/EMD-38541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38541 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38541 | HTTPS FTP |
-Related structure data
| Related structure data |  8xouMC  8xotC  8xowC  8xpmC  8xqbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38541.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38541.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.30654 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38541_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
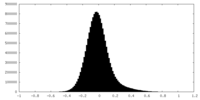
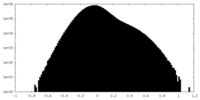
| Density Histograms |
-Half map: #2
| File | emd_38541_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
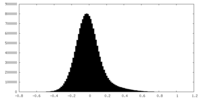
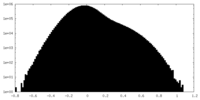
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage Lambda
| Entire | Name:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage Lambda
| Supramolecule | Name: Escherichia phage Lambda / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2681611 / Sci species name: Escherichia phage Lambda / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Portal protein B
| Macromolecule | Name: Portal protein B / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 59.529609 KDa |
| Recombinant expression | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Sequence | String: MKTPTIPTLL GPDGMTSLRE YAGYHGGGSG FGGQLRSWNP PSESVDAALL PNFTRGNARA DDLVRNNGYA ANAIQLHQDH IVGSFFRLS HRPSWRYLGI GEEEARAFSR EVEAAWKEFA EDDCCCIDVE RKRTFTMMIR EGVAMHAFNG ELFVQATWDT S SSRLFRTQ ...String: MKTPTIPTLL GPDGMTSLRE YAGYHGGGSG FGGQLRSWNP PSESVDAALL PNFTRGNARA DDLVRNNGYA ANAIQLHQDH IVGSFFRLS HRPSWRYLGI GEEEARAFSR EVEAAWKEFA EDDCCCIDVE RKRTFTMMIR EGVAMHAFNG ELFVQATWDT S SSRLFRTQ FRMVSPKRIS NPNNTGDSRN CRAGVQINDS GAALGYYVSE DGYPGWMPQK WTWIPRELPG GRASFIHVFE PV EDGQTRG ANVFYSVMEQ MKMLDTLQNT QLQSAIVKAM YAATIESELD TQSAMDFILG ANSQEQRERL TGWIGEIAAY YAA APVRLG GAKVPHLMPG DSLNLQTAQD TDNGYSVFEQ SLLRYIAAGL GVSYEQLSRN YAQMSYSTAR ASANESWAYF MGRR KFVAS RQASQMFLCW LEEAIVRRVV TLPSKARFSF QEARSAWGNC DWIGSGRMAI DGLKEVQEAV MLIEAGLSTY EKECA KRGD DYQEIFAQQV RETMERRAAG LKPPAWAAAA FESGLRQSTE EEKSDSRAA UniProtKB: Portal protein B |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 30 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 38.22916 KDa |
| Recombinant expression | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Sequence | String: MSMYTTAQLL AANEQKFKFD PLFLRLFFRE SYPFTTEKVY LSQIPGLVNM ALYVSPIVSG EVIRSRGGST SEFTPGYVKP KHEVNPQMT LRRLPDEDPQ NLADPAYRRR RIIMQNMRDE ELAIAQVEEM QAVSAVLKGK YTMTGEAFDP VEVDMGRSEE N NITQSGGT ...String: MSMYTTAQLL AANEQKFKFD PLFLRLFFRE SYPFTTEKVY LSQIPGLVNM ALYVSPIVSG EVIRSRGGST SEFTPGYVKP KHEVNPQMT LRRLPDEDPQ NLADPAYRRR RIIMQNMRDE ELAIAQVEEM QAVSAVLKGK YTMTGEAFDP VEVDMGRSEE N NITQSGGT EWSKRDKSTY DPTDDIEAYA LNASGVVNII VFDPKGWALF RSFKAVKEKL DTRRGSNSEL ETAVKDLGKA VS YKGMYGD VAIVVYSGQY VENGVKKNFL PDNTMVLGNT QARGLRTYGC IQDADAQREG INASARYPKN WVTTGDPARE FTM IQSAPL MLLADPDEFV SVQLA UniProtKB: Major capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)