+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Activation mechanism of GPR132 by compound NOX-6-7 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / GPR132 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of G2/M transition of mitotic cell cycle / Class A/1 (Rhodopsin-like receptors) / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity ...negative regulation of G2/M transition of mitotic cell cycle / Class A/1 (Rhodopsin-like receptors) / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / G1/S transition of mitotic cell cycle / positive regulation of cholesterol biosynthetic process / electron transport chain / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / iron ion binding / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / heme binding / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Wang JL / Ding JH / Sun JP / Yu X | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Metab / Year: 2023 Journal: Nat Metab / Year: 2023Title: Functional screening and rational design of compounds targeting GPR132 to treat diabetes. Authors: Jia-Le Wang / Xiao-Dong Dou / Jie Cheng / Ming-Xin Gao / Guo-Feng Xu / Wei Ding / Jin-Hui Ding / Yu Li / Si-Han Wang / Zhao-Wei Ji / Xin-Yi Zhao / Tong-Yu Huo / Cai-Fang Zhang / Ya-Meng Liu ...Authors: Jia-Le Wang / Xiao-Dong Dou / Jie Cheng / Ming-Xin Gao / Guo-Feng Xu / Wei Ding / Jin-Hui Ding / Yu Li / Si-Han Wang / Zhao-Wei Ji / Xin-Yi Zhao / Tong-Yu Huo / Cai-Fang Zhang / Ya-Meng Liu / Xue-Ying Sha / Jia-Rui Gao / Wen-Hui Zhang / Yong Hao / Cheng Zhang / Jin-Peng Sun / Ning Jiao / Xiao Yu /  Abstract: Chronic inflammation due to islet-residing macrophages plays key roles in the development of type 2 diabetes mellitus. By systematically profiling intra-islet lipid-transmembrane receptor signalling ...Chronic inflammation due to islet-residing macrophages plays key roles in the development of type 2 diabetes mellitus. By systematically profiling intra-islet lipid-transmembrane receptor signalling in islet-resident macrophages, we identified endogenous 9(S)-hydroxy-10,12-octadecadienoic acid-G-protein-coupled receptor 132 (GPR132)-Gi signalling as a significant contributor to islet macrophage reprogramming and found that GPR132 deficiency in macrophages reversed metabolic disorders in mice fed a high-fat diet. The cryo-electron microscopy structures of GPR132 bound with two endogenous agonists, N-palmitoylglycine and 9(S)-hydroxy-10,12-octadecadienoic acid, enabled us to rationally design both GPR132 agonists and antagonists with high potency and selectivity through stepwise translational approaches. We ultimately identified a selective GPR132 antagonist, NOX-6-18, that modulates macrophage reprogramming within pancreatic islets, decreases weight gain and enhances glucose metabolism in mice fed a high-fat diet. Our study not only illustrates that intra-islet lipid signalling contributes to islet macrophage reprogramming but also provides a broadly applicable strategy for the identification of important G-protein-coupled receptor targets in pathophysiological processes, followed by the rational design of therapeutic leads for refractory diseases such as diabetes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35044.map.gz emd_35044.map.gz | 11.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35044-v30.xml emd-35044-v30.xml emd-35044.xml emd-35044.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35044.png emd_35044.png | 108.6 KB | ||
| Filedesc metadata |  emd-35044.cif.gz emd-35044.cif.gz | 7.5 KB | ||
| Others |  emd_35044_half_map_1.map.gz emd_35044_half_map_1.map.gz emd_35044_half_map_2.map.gz emd_35044_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35044 http://ftp.pdbj.org/pub/emdb/structures/EMD-35044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35044 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35044 | HTTPS FTP |
-Related structure data
| Related structure data |  8hviMC  8hqeC  8hqmC  8hqnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35044.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35044.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
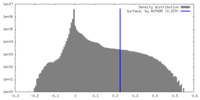
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35044_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
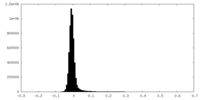
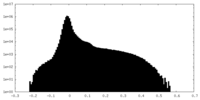
| Density Histograms |
-Half map: #1
| File | emd_35044_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
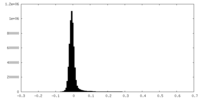
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the apo-GPR132-Gi
| Entire | Name: Cryo-EM structure of the apo-GPR132-Gi |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the apo-GPR132-Gi
| Supramolecule | Name: Cryo-EM structure of the apo-GPR132-Gi / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 kDa/nm |
-Macromolecule #1: Soluble cytochrome b562,Probable G-protein coupled receptor 132
| Macromolecule | Name: Soluble cytochrome b562,Probable G-protein coupled receptor 132 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.125316 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMCPMLLKNG YNGNATPVTT TAPWASLGLS A KTCNNVSF ...String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMCPMLLKNG YNGNATPVTT TAPWASLGLS A KTCNNVSF EESRIVLVVV YSAVCTLGVP ANCLTAWLAL LQVLQGNVLA VYLLCLALCE LLYTGTLPLW VIYIRNQHRW TL GLLACKV TAYIFFCNIY VSILFLCCIS CDRFVAVVYA LESRGRRRRR TAILISACIF ILVGIVHYPV FQTEDKETCF DML QMDSRI AGYYYARFTV GFAIPLSIIA FTNHRIFRSI KQSMGLSAAQ KAKVKHSAIA VVVIFLVCFA PYHLVLLVKA AAFS YYRGD RNAMCGLEER LYTASVVFLC LSTVNGVADP IIYVLATDHS RQEVSRIHKG WKEWSMKTDV TRLTHSRDTE ELQSP VALA DHYTFSRPVH PPGSPCPAKR LIEESC UniProtKB: Soluble cytochrome b562, Probable G-protein coupled receptor 132 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.48916 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MHHHHHHLEV LFQGPGSSQS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSQS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.610615 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGS |
-Macromolecule #6: 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofura...
| Macromolecule | Name: 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: NFI |
|---|---|
| Molecular weight | Theoretical: 351.353 Da |
| Chemical component information | 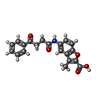 ChemComp-NFI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)