+Search query
-Structure paper
| Title | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome Complex Enabling Rational Design of Inhibitors. |
|---|---|
| Journal, issue, pages | J Am Chem Soc, Vol. 146, Issue 49, Page 33903-33913, Year 2024 |
| Publish date | Dec 11, 2024 |
 Authors Authors | Yu-Xia Wang / Ying Ye / Zhi-Wen Li / Guang-Rui Cui / Xing-Xing Shi / Ying Dong / Jia-Jia Jiang / Jia-Yue Sun / Ze-Wei Guan / Nan Zhang / Qiong-You Wu / Fan Wang / Xiao-Lei Zhu / Guang-Fu Yang /  |
| PubMed Abstract | Cytochrome (complex III) represents a significant target for the discovery of both drugs and fungicides. Metyltetraprole (MET) is commonly classified as a quinone site inhibitor (QI) that combats ...Cytochrome (complex III) represents a significant target for the discovery of both drugs and fungicides. Metyltetraprole (MET) is commonly classified as a quinone site inhibitor (QI) that combats the G143A mutated isolate, which confers high resistance to strobilurin fungicides such as pyraclostrobin (PYR). The binding mode and antiresistance mechanism of MET remain unclear. Here, we determined the high-resolution structures of inhibitor-bound complex III (MET, 2.52 Å; PYR, 2.42 Å) and inhibitor-bound porcine complex III (MET, 2.53 Å; PYR, 2,37 Å) by cryo-electron microscopy. The distinct binding modes of MET and PYR were observed for the first time. Notably, the MET exhibited different binding modes in the two species. In , the binding site of MET was the same as PYR, serving as a -type inhibitor of the Q site. However, in porcine, MET acted as a dual-target inhibitor of both Q and Q. Based on the structural insights, a novel inhibitor (YF23694) was discovered and demonstrated excellent fungicidal activity against downy mildew and powdery mildew fungi. This work provides a new starting point for the design of the next generation of inhibitors to overcome the resistance. |
 External links External links |  J Am Chem Soc / J Am Chem Soc /  PubMed:39601138 PubMed:39601138 |
| Methods | EM (single particle) |
| Resolution | 2.37 - 2.74 Å |
| Structure data | EMDB-39291, PDB-8yhq: EMDB-39323, PDB-8yin: EMDB-60256, PDB-8zmt: EMDB-60317, PDB-8zos: EMDB-60320, PDB-8zow: EMDB-60323, PDB-8zp0: |
| Chemicals |  ChemComp-6PH: 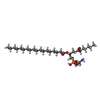 ChemComp-9PE:  ChemComp-HEM:  ChemComp-8PE:  ChemComp-CN5:  ChemComp-UQ6:  PDB-1d6k:  ChemComp-7PH: 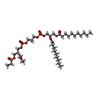 ChemComp-CN3:  PDB-1d6o:  PDB-1d6p:  ChemComp-FES:  ChemComp-PEE:  ChemComp-HEC:  ChemComp-CDL: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Complex / mitochondria / ELECTRON TRANSPORT / PROTEIN BINDING / Pyraclostrobin / Metyltetraprole / Inhibitor |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers

















