-Search query
-Search result
Showing all 41 items for (author: skehel & jm)

EMDB-17114: 
CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2

EMDB-17115: 
CAND1-CUL1-RBX1-DCNL1

EMDB-17116: 
CAND1-CUL1-RBX1-SKP1-SKP2-DCNL1

EMDB-17117: 
Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2

EMDB-17124: 
CAND1-CUL1-RBX1
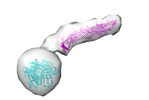
EMDB-15604: 
ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation
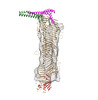
EMDB-15605: 
Low resolution 3D reconstruction of ATG2A from cryo-EM
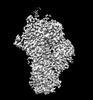
EMDB-14710: 
Polymerase module of CPF in complex with Mpe1 and a pre-cleaved CYC1 RNA

EMDB-14711: 
Polymerase module of yeast CPF in complex with the yPIM of Cft2

EMDB-14712: 
Polymerase module of yeast CPF in complex with Mpe1, the yPIM of Cft2 and the pre-cleaved CYC1 RNA

EMDB-13233: 
Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2

EMDB-12957: 
Cryo-EM structure of the human R2TP-TTT complex

EMDB-12979: 
Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex

EMDB-11220: 
Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class.

EMDB-11221: 
Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class.

EMDB-11240: 
Cryo-EM structure of the E.coli HTL-BAm complex

EMDB-11302: 
Structure of a human 48S translational initiation complex

EMDB-10769: 
Structure of a human 48S translational initiation complex - eIF3

EMDB-10772: 
Structure of a human 48S translational initiation complex - head

EMDB-10773: 
Structure of a human 48S translational initiation complex - eIF3bgi

EMDB-10774: 
Structure of a human 48S translational initiation complex - eIF2-TC

EMDB-10775: 
Structure of a human 48S translational initiation complex - 40S body

EMDB-10277: 
Cryo-EM structure of human SERINC5 in LMNG micelles, bound to Fab.

EMDB-10279: 
CryoEM structure of SERINC from Drosophila melanogaster

EMDB-10290: 
Structure of Fanconi anaemia core complex (consensus map)

EMDB-10291: 
Structure of the Fanconi Anaemia core complex (focussed map for top region)

EMDB-10292: 
Structure of the Fanconi Anaemia core complex (focussed map for middle region)

EMDB-10293: 
Structure of the Fanconi Anaemia core complex (focussed map for base region)

EMDB-10294: 
Structure of the Fanconi anaemia core subcomplex

EMDB-10186: 
Structure of the ESX-3 core complex

EMDB-10187: 
Structure of protomer 1 of the ESX-3 core complex

EMDB-10188: 
Structure of EccB3 dimer from the ESX-3 core complex

EMDB-10189: 
ESX-3 core complex centred at the cytoplasmic region, conformation 1.

EMDB-10190: 
ESX-3 core complex centred at the cytoplasmic region, conformation 2.

EMDB-10191: 
Structure of protomer 2 of the ESX-3 core complex

EMDB-4287: 
Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3.

EMDB-4289: 
Human R2TP complex-C3symmetry

EMDB-4290: 
Human R2TP complex-subcomplex1

EMDB-4291: 
Human R2TP complex-subgroup2

EMDB-3677: 
Structure of the heterohexameric yeast Rvb1-Rvb2 complex

EMDB-3678: 
Structure of the yeast R2TP complex
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
