+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10186 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the ESX-3 core complex | ||||||||||||||||||
 Map data Map data | Structure of the ESX-3 core dimer | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Type VII Secretion System ESX-3 secretion system T7SS ESX-3 Mycobacterium smegmatis / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides / hydrolase activity / ATP hydrolysis activity / DNA binding / extracellular region / ATP binding / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) / Mycolicibacterium smegmatis MC2 155 (bacteria) /  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) | ||||||||||||||||||
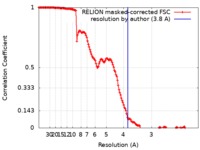
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Rivera-Calzada A / Famelis N / Geibel S / Llorca O | ||||||||||||||||||
| Funding support |  Spain, Spain,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Architecture of the mycobacterial type VII secretion system. Authors: Nikolaos Famelis / Angel Rivera-Calzada / Gianluca Degliesposti / Maria Wingender / Nicole Mietrach / J Mark Skehel / Rafael Fernandez-Leiro / Bettina Böttcher / Andreas Schlosser / Oscar ...Authors: Nikolaos Famelis / Angel Rivera-Calzada / Gianluca Degliesposti / Maria Wingender / Nicole Mietrach / J Mark Skehel / Rafael Fernandez-Leiro / Bettina Böttcher / Andreas Schlosser / Oscar Llorca / Sebastian Geibel /    Abstract: Host infection by pathogenic mycobacteria, such as Mycobacterium tuberculosis, is facilitated by virulence factors that are secreted by type VII secretion systems. A molecular understanding of the ...Host infection by pathogenic mycobacteria, such as Mycobacterium tuberculosis, is facilitated by virulence factors that are secreted by type VII secretion systems. A molecular understanding of the type VII secretion mechanism has been hampered owing to a lack of three-dimensional structures of the fully assembled secretion apparatus. Here we report the cryo-electron microscopy structure of a membrane-embedded core complex of the ESX-3/type VII secretion system from Mycobacterium smegmatis. The core of the ESX-3 secretion machine consists of four protein components-EccB3, EccC3, EccD3 and EccE3, in a 1:1:2:1 stoichiometry-which form two identical protomers. The EccC3 coupling protein comprises a flexible array of four ATPase domains, which are linked to the membrane through a stalk domain. The domain of unknown function (DUF) adjacent to the stalk is identified as an ATPase domain that is essential for secretion. EccB3 is predominantly periplasmatic, but a small segment crosses the membrane and contacts the stalk domain. This suggests that conformational changes in the stalk domain-triggered by substrate binding at the distal end of EccC3 and subsequent ATP hydrolysis in the DUF-could be coupled to substrate secretion to the periplasm. Our results reveal that the architecture of type VII secretion systems differs markedly from that of other known secretion machines, and provide a structural understanding of these systems that will be useful for the design of antimicrobial strategies that target bacterial virulence. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10186.map.gz emd_10186.map.gz | 256.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10186-v30.xml emd-10186-v30.xml emd-10186.xml emd-10186.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10186_fsc.xml emd_10186_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10186.png emd_10186.png | 66.1 KB | ||
| Filedesc metadata |  emd-10186.cif.gz emd-10186.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10186 http://ftp.pdbj.org/pub/emdb/structures/EMD-10186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10186 | HTTPS FTP |
-Related structure data
| Related structure data |  6sgwMC  6sgxC  6sgyC  6sgzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10186.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10186.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the ESX-3 core dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : ESX-3 core complex
| Entire | Name: ESX-3 core complex |
|---|---|
| Components |
|
-Supramolecule #1: ESX-3 core complex
| Supramolecule | Name: ESX-3 core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The sample consists of four protein components, EccB3:EccC3:EccD3:EccE3 in a 1:1:2:1 stoichiometry Molecular weight of the complex without the amphipol micelle: 0.65 MDa |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 650 KDa |
-Macromolecule #1: ESX-3 secretion system ATPase EccB3
| Macromolecule | Name: ESX-3 secretion system ATPase EccB3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 9.407883 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: DRRSFSSRTP VNENPDGVQY RRGFVTRHQV SGWRFVMRRI ASGVALHDTR MLVDPLRTQS RAVLTGALIL VTGLVGCFIF SLF UniProtKB: ESX-3 secretion system ATPase EccB3 |
-Macromolecule #2: ESX-3 secretion system protein EccD3
| Macromolecule | Name: ESX-3 secretion system protein EccD3 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 47.114125 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: PIVRVAVLAA GDDGGRLTEM ALPSELPLRE ILPAVQRIVQ PARENDGAAD PAAAPNPVRL SLAPIGGAPF SLDATLDTVG VVDGDLLAL QAVPSGPPAP RIVEDIADAA VIFSEARRRQ WGPTHIARGA ALALIGLILV GTGLSVAHRV ITGDLLGQFI V SGIALATV ...String: PIVRVAVLAA GDDGGRLTEM ALPSELPLRE ILPAVQRIVQ PARENDGAAD PAAAPNPVRL SLAPIGGAPF SLDATLDTVG VVDGDLLAL QAVPSGPPAP RIVEDIADAA VIFSEARRRQ WGPTHIARGA ALALIGLILV GTGLSVAHRV ITGDLLGQFI V SGIALATV IAALAVRNRS AVLATSLAVT ALVPVAAAFA LGVPGDFGAP NVLLAAAGVA AWSLISMAGS PDDRGIAVFT AT AVTGVGV LLVAGAASLW VISSDVIGCA LVLLGLIVTV QAAQLSAMWA RFPLPVIPAP GDPTPAARPL SVLADLPRRV RVS QAHQTG VIAAGVLLGV AGSVALVSSA NASPWAWYIV VAAAAGAALR ARVWDSAACK AWLLGHSYLL AVALLVAFVI GDRY QAALW ALAALAVLVL VWIVAALNPK IASPDTYSLP MRRMVGFLAT GLDASLIPVM ALLVGLFSLV UniProtKB: ESX-3 secretion system protein EccD3 |
-Macromolecule #3: ESX-3 secretion system protein EccC3
| Macromolecule | Name: ESX-3 secretion system protein EccC3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 44.653078 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: SRLIFEHQRR LTPPTTRKGT ITIEPPPQLP RVVPPSLLRR VLPFLIVILI VGMIVALFAT GMRLISPTML FFPFVLLLAA TALYRGGDN KMRTEEVDAE RADYLRYLSV VRDNVRAHAA EQRAALEWSH PEPEVLATIP GTRRQWERDP RDRDFLVLRA G RHDVPLDA ...String: SRLIFEHQRR LTPPTTRKGT ITIEPPPQLP RVVPPSLLRR VLPFLIVILI VGMIVALFAT GMRLISPTML FFPFVLLLAA TALYRGGDN KMRTEEVDAE RADYLRYLSV VRDNVRAHAA EQRAALEWSH PEPEVLATIP GTRRQWERDP RDRDFLVLRA G RHDVPLDA ALKVKDTADE IDLEPVAHSA LRGLLDVQRT VRDAPTGLDV AKLARITVIG EADEARAAIR AWIAQAVTWH DP TMLGVAL AAPDLESGDW SWLKWLPHVD VPNEADGVGP ARYLTTSTAE LRERLAPALA DRPLFPAESG AALKHLLVVL DDP DADPDD IARKPGLTGV TVIHRTTELP NREQYPDPER PILRVADGRI ERWQVGGWQP CVDVADAMSA AEAAHIARRL SRWD S UniProtKB: ESX-3 secretion system protein EccC3 |
-Macromolecule #4: ESX-3 secretion system protein EccE3
| Macromolecule | Name: ESX-3 secretion system protein EccE3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 30.813355 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MTARIALASL FVVAAVLAQP WQTTTQRWVL GVSIAAVIVL LAWWKGMFLT TRIGRALAMV RRNRAEDTVE TDAHRATVVL RVDPAAPAQ LPVVVGYLDR YGITCDKVRI THRDAGGTRR SWISLTVDAV DNLAALQARS ARIPLQDTTE VVGRRLADHL R EQGWTVTV ...String: MTARIALASL FVVAAVLAQP WQTTTQRWVL GVSIAAVIVL LAWWKGMFLT TRIGRALAMV RRNRAEDTVE TDAHRATVVL RVDPAAPAQ LPVVVGYLDR YGITCDKVRI THRDAGGTRR SWISLTVDAV DNLAALQARS ARIPLQDTTE VVGRRLADHL R EQGWTVTV VEGVDTPLPV SGKETWRGVA DDAGVVAAYR VKVDDRLDEV LAEIGHLPAE ETWTALEFTG SPAEPLLTVC AA VRTSDRP AAKAPLAGLT PARGRHRPAL AALNPLSTER LDGTAVPL UniProtKB: ESX-3 secretion system protein EccE3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 30 mM Hepes pH 8.0, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time: 3s Blotting force: -10. |
| Details | bound to Amphipol A8-35 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 11903 / Average electron dose: 50.0 e/Å2 Details: Images acquired as 55 frames movies at a calibrated magnification of 1.0635 Angs/px |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 76.45 / Target criteria: Map to model correlation coefficient |
|---|---|
| Output model |  PDB-6sgw: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)