+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11240 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the E.coli HTL-BAm complex | |||||||||||||||
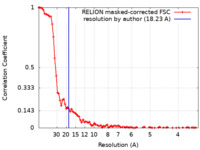
 Map data Map data | Cryo-EM of the E. coli HTL-BAM complex in detergent. 18,23A resolution. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 18.23 Å | |||||||||||||||
 Authors Authors | Alvira S / Collinson S | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Inter-membrane association of the Sec and BAM translocons for bacterial outer-membrane biogenesis. Authors: Sara Alvira / Daniel W Watkins / Luca Troman / William J Allen / James S Lorriman / Gianluca Degliesposti / Eli J Cohen / Morgan Beeby / Bertram Daum / Vicki Am Gold / J Mark Skehel / Ian Collinson /  Abstract: The outer-membrane of Gram-negative bacteria is critical for surface adhesion, pathogenicity, antibiotic resistance and survival. The major constituent - hydrophobic β-barrel uter-embrane roteins ...The outer-membrane of Gram-negative bacteria is critical for surface adhesion, pathogenicity, antibiotic resistance and survival. The major constituent - hydrophobic β-barrel uter-embrane roteins (OMPs) - are first secreted across the inner-membrane through the Sec-translocon for delivery to periplasmic chaperones, for example SurA, which prevent aggregation. OMPs are then offloaded to the β-arrel ssembly achinery (BAM) in the outer-membrane for insertion and folding. We show the olo-ransocon (HTL) - an assembly of the protein-channel core-complex SecYEG, the ancillary sub-complex SecDF, and the membrane 'insertase' YidC - contacts BAM through periplasmic domains of SecDF and YidC, ensuring efficient OMP maturation. Furthermore, the proton-motive force (PMF) across the inner-membrane acts at distinct stages of protein secretion: (1) SecA-driven translocation through SecYEG and (2) communication of conformational changes via SecDF across the periplasm to BAM. The latter presumably drives efficient passage of OMPs. These interactions provide insights of inter-membrane organisation and communication, the importance of which is becoming increasingly apparent. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11240.map.gz emd_11240.map.gz | 54.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11240-v30.xml emd-11240-v30.xml emd-11240.xml emd-11240.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11240_fsc.xml emd_11240_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_11240.png emd_11240.png | 72.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11240 http://ftp.pdbj.org/pub/emdb/structures/EMD-11240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11240 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11240.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11240.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM of the E. coli HTL-BAM complex in detergent. 18,23A resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.75 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli inner membrane holotranslocon in complex with the outer m...
| Entire | Name: E. coli inner membrane holotranslocon in complex with the outer membrane complex BAM |
|---|---|
| Components |
|
-Supramolecule #1: E. coli inner membrane holotranslocon in complex with the outer m...
| Supramolecule | Name: E. coli inner membrane holotranslocon in complex with the outer membrane complex BAM type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 205 KDa |
-Supramolecule #2: E coli HTL
| Supramolecule | Name: E coli HTL / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: E. coli BAM
| Supramolecule | Name: E. coli BAM / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 20mM HEPES, pH 8.0 250 mM NaCl, 0.03% (w/v) DDM/0.003% (w/v) CL |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.02 nm / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Average exposure time: 20.0 sec. / Average electron dose: 1.127 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 79000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)