[English] 日本語
 Yorodumi
Yorodumi- EMDB-3825: Three-dimensional cryo-EM density map of paired C2S2M PSII-LHCII ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3825 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional cryo-EM density map of paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativum | |||||||||
 Map data Map data | Three-dimensional cryo-EM density map of paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativum | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pisum sativum (garden pea) Pisum sativum (garden pea) | |||||||||
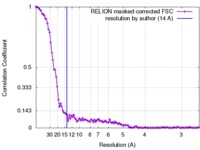
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | Pagliano C | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2017 Journal: Sci Rep / Year: 2017Title: Pea PSII-LHCII supercomplexes form pairs by making connections across the stromal gap. Authors: Pascal Albanese / Roberto Melero / Benjamin D Engel / Alessandro Grinzato / Paola Berto / Marcello Manfredi / Angelica Chiodoni / Javier Vargas / Carlos Óscar Sánchez Sorzano / Emilio ...Authors: Pascal Albanese / Roberto Melero / Benjamin D Engel / Alessandro Grinzato / Paola Berto / Marcello Manfredi / Angelica Chiodoni / Javier Vargas / Carlos Óscar Sánchez Sorzano / Emilio Marengo / Guido Saracco / Giuseppe Zanotti / Jose-Maria Carazo / Cristina Pagliano /    Abstract: In higher plant thylakoids, the heterogeneous distribution of photosynthetic protein complexes is a determinant for the formation of grana, stacks of membrane discs that are densely populated with ...In higher plant thylakoids, the heterogeneous distribution of photosynthetic protein complexes is a determinant for the formation of grana, stacks of membrane discs that are densely populated with Photosystem II (PSII) and its light harvesting complex (LHCII). PSII associates with LHCII to form the PSII-LHCII supercomplex, a crucial component for solar energy conversion. Here, we report a biochemical, structural and functional characterization of pairs of PSII-LHCII supercomplexes, which were isolated under physiologically-relevant cation concentrations. Using single-particle cryo-electron microscopy, we determined the three-dimensional structure of paired CSM PSII-LHCII supercomplexes at 14 Å resolution. The two supercomplexes interact on their stromal sides through a specific overlap between apposing LHCII trimers and via physical connections that span the stromal gap, one of which is likely formed by interactions between the N-terminal loops of two Lhcb4 monomeric LHCII subunits. Fast chlorophyll fluorescence induction analysis showed that paired PSII-LHCII supercomplexes are energetically coupled. Molecular dynamics simulations revealed that additional flexible physical connections may form between the apposing LHCII trimers of paired PSII-LHCII supercomplexes in appressed thylakoid membranes. Our findings provide new insights into how interactions between pairs of PSII-LHCII supercomplexes can link adjacent thylakoids to mediate the stacking of grana membranes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3825.map.gz emd_3825.map.gz | 235.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3825-v30.xml emd-3825-v30.xml emd-3825.xml emd-3825.xml | 10.6 KB 10.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3825_fsc.xml emd_3825_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3825.png emd_3825.png | 65.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3825 http://ftp.pdbj.org/pub/emdb/structures/EMD-3825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3825 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10198 (Title: Paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativum EMPIAR-10198 (Title: Paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativumData size: 433.1 / Data #1: 2.outputMicrographs [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3825.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3825.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Three-dimensional cryo-EM density map of paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativum | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : paired PSII-LHCII supercomplexes from pea plants
| Entire | Name: paired PSII-LHCII supercomplexes from pea plants |
|---|---|
| Components |
|
-Supramolecule #1: paired PSII-LHCII supercomplexes from pea plants
| Supramolecule | Name: paired PSII-LHCII supercomplexes from pea plants / type: complex / ID: 1 / Parent: 0 Details: paired PSII-LHCII supercomplexes connected across the stromal gap |
|---|---|
| Source (natural) | Organism:  Pisum sativum (garden pea) / Strain: Palladio nano Pisum sativum (garden pea) / Strain: Palladio nano |
| Molecular weight | Theoretical: 2.75 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 5.7 / Component - Concentration: 25.0 mM / Component - Formula: C6H13NO4S / Component - Name: MES / Details: fresh solution |
| Grid | Model: Quantifoil lacey carbon / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK III / Details: blot for 4 seconds befoere plunging. |
| Details | Specimen concentration (mg/mL) is given in terms of Chlorophyll |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 6834 / Average exposure time: 1.5 sec. / Average electron dose: 7.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)