[English] 日本語
 Yorodumi
Yorodumi- EMDB-10277: Cryo-EM structure of human SERINC5 in LMNG micelles, bound to Fab. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10277 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human SERINC5 in LMNG micelles, bound to Fab. | |||||||||
 Map data Map data | sharpened EM map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
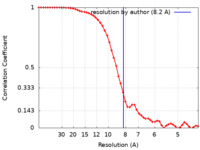
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | |||||||||
 Authors Authors | Rosa A / Pye VE / Nans A / Cherepanov P | |||||||||
| Funding support |  United States, United States,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Authors: Valerie E Pye / Annachiara Rosa / Cinzia Bertelli / Weston B Struwe / Sarah L Maslen / Robin Corey / Idlir Liko / Mark Hassall / Giada Mattiuzzo / Allison Ballandras-Colas / Andrea Nans / ...Authors: Valerie E Pye / Annachiara Rosa / Cinzia Bertelli / Weston B Struwe / Sarah L Maslen / Robin Corey / Idlir Liko / Mark Hassall / Giada Mattiuzzo / Allison Ballandras-Colas / Andrea Nans / Yasuhiro Takeuchi / Phillip J Stansfeld / J Mark Skehel / Carol V Robinson / Massimo Pizzato / Peter Cherepanov /   Abstract: The human integral membrane protein SERINC5 potently restricts HIV-1 infectivity and sensitizes the virus to antibody-mediated neutralization. Here, using cryo-EM, we determine the structures of ...The human integral membrane protein SERINC5 potently restricts HIV-1 infectivity and sensitizes the virus to antibody-mediated neutralization. Here, using cryo-EM, we determine the structures of human SERINC5 and its orthologue from Drosophila melanogaster at subnanometer and near-atomic resolution, respectively. The structures reveal a novel fold comprised of ten transmembrane helices organized into two subdomains and bisected by a long diagonal helix. A lipid binding groove and clusters of conserved residues highlight potential functional sites. A structure-based mutagenesis scan identified surface-exposed regions and the interface between the subdomains of SERINC5 as critical for HIV-1-restriction activity. The same regions are also important for viral sensitization to neutralizing antibodies, directly linking the antiviral activity of SERINC5 with remodeling of the HIV-1 envelope glycoprotein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10277.map.gz emd_10277.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10277-v30.xml emd-10277-v30.xml emd-10277.xml emd-10277.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10277_fsc.xml emd_10277_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_10277.png emd_10277.png | 125.3 KB | ||
| Others |  emd_10277_additional.map.gz emd_10277_additional.map.gz emd_10277_additional_1.map.gz emd_10277_additional_1.map.gz emd_10277_half_map_1.map.gz emd_10277_half_map_1.map.gz emd_10277_half_map_2.map.gz emd_10277_half_map_2.map.gz | 7.8 MB 7.8 MB 7.9 MB 7.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10277 http://ftp.pdbj.org/pub/emdb/structures/EMD-10277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10277 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10277.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10277.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened EM map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unsharpened EM map
| File | emd_10277_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
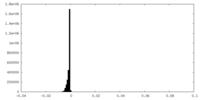
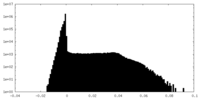
| Density Histograms |
-Additional map: unsharpened EM map
| File | emd_10277_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
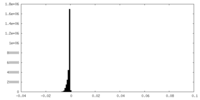
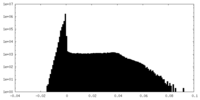
| Density Histograms |
-Half map: EM half map 2
| File | emd_10277_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EM half map 1
| File | emd_10277_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : complex of SERINC5 with FAB bound to ECL4 in LMNG micelles
| Entire | Name: complex of SERINC5 with FAB bound to ECL4 in LMNG micelles |
|---|---|
| Components |
|
-Supramolecule #1: complex of SERINC5 with FAB bound to ECL4 in LMNG micelles
| Supramolecule | Name: complex of SERINC5 with FAB bound to ECL4 in LMNG micelles type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Macromolecule #1: SERINC5
| Macromolecule | Name: SERINC5 / type: protein_or_peptide / ID: 1 Details: Full-length, wild type human SERINC5 with C-terminal TwinStrep tag. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: msaqccagql acccgsagcs lccdccprir qslstrfmya lyfilvvvlc cimmsttvah kmkehipffe dmckgikagd tceklvgysa vyrvcfgmac fffifclltl kinnskscra hihngfwffk llllgamcsg affipdqdtf lnawryvgav ggflfigiql ...String: msaqccagql acccgsagcs lccdccprir qslstrfmya lyfilvvvlc cimmsttvah kmkehipffe dmckgikagd tceklvgysa vyrvcfgmac fffifclltl kinnskscra hihngfwffk llllgamcsg affipdqdtf lnawryvgav ggflfigiql lllvefahkw nknwtagtas nklwyaslal vtlimysiat gglvlmavfy tqkdscmenk illgvngglc llislvaisp wvqnrqphsg llqsgviscy vtyltfsals skpaevvlde hgknvticvp dfgqdlyrde nlvtilgtsl ligcilyscl tsttrsssda lqgryaapel eiarccfcfs pggedteeqq pgkegprviy dekkgtvyiy syfhfvffla slyvmmtvtn wfnyesanie sffsgswsif wvkmascwic vllylctlva plccptrefs vgssglevlf qgpgsggsaw shpqfekggg sgggsggsaw shpqfek |
-Macromolecule #2: FAB (antigen binding fragment from a monoclonal antibody)
| Macromolecule | Name: FAB (antigen binding fragment from a monoclonal antibody) type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: amino acid sequence unknown |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Details: No pre-treatment | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | The complex was assembled using recombinant SERINC5 in LMNG/PS and FAB and was purified by size exclusion chromatograhy immediately prior to vitrification on holey carbon EM grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 7165 / Average electron dose: 33.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -4.0 µm / Nominal defocus min: -1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)