[English] 日本語
 Yorodumi
Yorodumi- EMDB-15604: ATG9A and ATG2A form a heteromeric complex essential for autophag... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
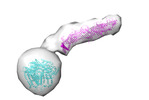
| Title | ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Lipid transfer / lipid scramblase / membrane protein / protein complex / LIPID TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphagophore / lipid transfer activity / organelle membrane contact site / glycophagy / negative regulation of macrophage cytokine production / phospholipid scramblase activity / protein localization to Golgi apparatus / programmed necrotic cell death / positive regulation of autophagosome assembly / protein localization to phagophore assembly site ...phagophore / lipid transfer activity / organelle membrane contact site / glycophagy / negative regulation of macrophage cytokine production / phospholipid scramblase activity / protein localization to Golgi apparatus / programmed necrotic cell death / positive regulation of autophagosome assembly / protein localization to phagophore assembly site / phagophore assembly site membrane / pexophagy / autophagy of mitochondrion / piecemeal microautophagy of the nucleus / phosphatidylinositol-3-phosphate binding / negative regulation of interferon-beta production / bone morphogenesis / phagophore assembly site / reticulophagy / Macroautophagy / autophagosome assembly / mitophagy / protein-membrane adaptor activity / lipid droplet / positive regulation of autophagy / autophagosome / synaptic membrane / PINK1-PRKN Mediated Mitophagy / trans-Golgi network / recycling endosome / mitochondrial membrane / recycling endosome membrane / late endosome / late endosome membrane / endosome / intracellular membrane-bounded organelle / Golgi membrane / innate immune response / endoplasmic reticulum membrane / Golgi apparatus / mitochondrion / membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 32.89 Å | ||||||||||||
 Authors Authors | Chiduza GN / van Vliet AR / De Tito S / Punch EK / Tooze SA | ||||||||||||
| Funding support |  United Kingdom, European Union, 3 items United Kingdom, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Authors: Alexander R van Vliet / George N Chiduza / Sarah L Maslen / Valerie E Pye / Dhira Joshi / Stefano De Tito / Harold B J Jefferies / Evangelos Christodoulou / Chloë Roustan / Emma Punch / ...Authors: Alexander R van Vliet / George N Chiduza / Sarah L Maslen / Valerie E Pye / Dhira Joshi / Stefano De Tito / Harold B J Jefferies / Evangelos Christodoulou / Chloë Roustan / Emma Punch / Javier H Hervás / Nicola O'Reilly / J Mark Skehel / Peter Cherepanov / Sharon A Tooze /   Abstract: ATG9A and ATG2A are essential core members of the autophagy machinery. ATG9A is a lipid scramblase that allows equilibration of lipids across a membrane bilayer, whereas ATG2A facilitates lipid flow ...ATG9A and ATG2A are essential core members of the autophagy machinery. ATG9A is a lipid scramblase that allows equilibration of lipids across a membrane bilayer, whereas ATG2A facilitates lipid flow between tethered membranes. Although both have been functionally linked during the formation of autophagosomes, the molecular details and consequences of their interaction remain unclear. By combining data from peptide arrays, crosslinking, and hydrogen-deuterium exchange mass spectrometry together with cryoelectron microscopy, we propose a molecular model of the ATG9A-2A complex. Using this integrative structure modeling approach, we identify several interfaces mediating ATG9A-2A interaction that would allow a direct transfer of lipids from ATG2A into the lipid-binding perpendicular branch of ATG9A. Mutational analyses combined with functional activity assays demonstrate their importance for autophagy, thereby shedding light on this protein complex at the heart of autophagy. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15604.map.gz emd_15604.map.gz | 446.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15604-v30.xml emd-15604-v30.xml emd-15604.xml emd-15604.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15604_fsc.xml emd_15604_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15604.png emd_15604.png | 43.4 KB | ||
| Masks |  emd_15604_msk_1.map emd_15604_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15604.cif.gz emd-15604.cif.gz | 6.9 KB | ||
| Others |  emd_15604_half_map_1.map.gz emd_15604_half_map_1.map.gz emd_15604_half_map_2.map.gz emd_15604_half_map_2.map.gz | 21.2 MB 21.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15604 http://ftp.pdbj.org/pub/emdb/structures/EMD-15604 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15604 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15604 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15604.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15604.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.4 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15604_msk_1.map emd_15604_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15604_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15604_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterotetrameric complex of autophagy related proteins ATG9A and ATG2A
| Entire | Name: Heterotetrameric complex of autophagy related proteins ATG9A and ATG2A |
|---|---|
| Components |
|
-Supramolecule #1: Heterotetrameric complex of autophagy related proteins ATG9A and ATG2A
| Supramolecule | Name: Heterotetrameric complex of autophagy related proteins ATG9A and ATG2A type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Autophagy-related protein 9A
| Macromolecule | Name: Autophagy-related protein 9A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens environmental sample (environmental samples) Homo sapiens environmental sample (environmental samples) |
| Sequence | String: MDYKDDDDKD YKDDDDKDYK DDDDKHHHHH HENLYFQGMA QFDTEYQRLE ASYSDSPPGE EDLLVHVAEG SKSPWHHIEN LDLFFSRVYN LHQKNGFT C MLIGEIFELM QFLFVVAFTT FLVSCVDYDI LFANKMVNHS LHPTEPVKVT LPDAFLPAQ VCSARIQENG ...String: MDYKDDDDKD YKDDDDKDYK DDDDKHHHHH HENLYFQGMA QFDTEYQRLE ASYSDSPPGE EDLLVHVAEG SKSPWHHIEN LDLFFSRVYN LHQKNGFT C MLIGEIFELM QFLFVVAFTT FLVSCVDYDI LFANKMVNHS LHPTEPVKVT LPDAFLPAQ VCSARIQENG SLITILVIAG VFWIHRLIKF IYNICCYWEI HSFYLHALRI PMSALPYCTW QEVQARIVQ TQKEHQICIH KRELTELDIY HRILRFQNYM VALVNKSLLP LRFRLPGLGE A VFFTRGLK YNFELILFWG PGSLFLNEWS LKAEYKRGGQ RLELAQRLSN RILWIGIANF LL CPLILIW QILYAFFSYA EVLKREPGAL GARCWSLYGR CYLRHFNELE HELQSRLNRG YKP ASKYMN CFLSPLLTLL AKNGAFFAGS ILAVLIALTI YDEDVLAVEH VLTTVTLLGV TVTV CRSFI PDQHMVFCPE QLLRVILAHI HYMPDHWQGN AHRSQTRDEF AQLFQYKAVF ILEEL LSPI VTPLILIFCL RPRALEIIDF FRNFTVEVVG VGDTCSFAQM DVRQHGHPQW LSAGQT EAS VYQQAEDGKT ELSLMHFAIT NPGWQPPRES TAFLGFLKEQ VQRDGAAASL AQGGLLP EN ALFTSIQSLQ SESEPLSLIA NVVAGSSCRG PPLPRDLQGS RHRAEVASAL RSFSPLQP G QAPTGRAHST MTGSGVDART ASSGSSVWEG QLQSLVLSEY ASTEMSLHAL YMHQLHKQQ AQAEPERHVW HRRESDESGE SAPDEGGEGA RAPQSIPRSA SYPCAAPRPG APETTALHGG FQRRYGGIT DPGTVPRVPS HFSRLPLGGW AEDGQSASRH PEPVPEEGSE DELPPQVHKV UniProtKB: Autophagy-related protein 9A |
-Macromolecule #2: Autophagy-related protein 2 homolog A
| Macromolecule | Name: Autophagy-related protein 2 homolog A / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKD YKDDDDKDYK DDDDKGSGAG AGAGAILNSR VSGLRSRMSR WLWPWSNCVK ERVCRYLLHH YLGHFFQEHL SLDQLSLDLY KGSVALRDIH LEIWSVNEVL ESMESPLELV EGFVGSIEVA VPWAALLTDH CTVRVSGLQL TLQPRRGPAP GAADSQSWAS ...String: MDYKDDDDKD YKDDDDKDYK DDDDKGSGAG AGAGAILNSR VSGLRSRMSR WLWPWSNCVK ERVCRYLLHH YLGHFFQEHL SLDQLSLDLY KGSVALRDIH LEIWSVNEVL ESMESPLELV EGFVGSIEVA VPWAALLTDH CTVRVSGLQL TLQPRRGPAP GAADSQSWAS CMTTSLQLAQ ECLRDGLPEP SEPPQPLEGL EMFAQTIETV LRRIKVTFLD TVVRVEHSPG DGERGVAVEV RVQRLEYCDE AVRDPSQAPP VDVHQPPAFL HKLLQLAGVR LHYEELPAQE EPPEPPLQIG SCSGYMELMV KLKQNEAFPG PKLEVAGQLG SLHLLLTPRQ LQQLQELLSA VSLTDHEGLA DKLNKSRPLG AEDLWLIEQD LNQQLQAGAV AEPLSPDPLT NPLLNLDNTD LFFSMAGLTS SVASALSELS LSDVDLASSV RSDMASRRLS AQAHPAGKMA PNPLLDTMRP DSLLKMTLGG VTLTLLQTSA PSSGPPDLAT HFFTEFDATK DGPFGSRDFH HLRPRFQRAC PCSHVRLTGT AVQLSWELRT GSRGRRTTSM EVHFGQLEVL ECLWPRGTSE PEYTEILTFP GTLGSQASAR PCAHLRHTQI LRRVPKSRPR RSVACHCHSE LALDLANFQA DVELGALDRL AALLRLATVP AEPPAGLLTE PLPAMEQQTV FRLSAPRATL RLRFPIADLR PEPDPWAGQA VRAEQLRLEL SEPQFRSELS SGPGPPVPTH LELTCSDLHG IYEDGGKPPV PCLRVSKALD PKSTGRKYFL PQVVVTVNPQ SSSTQWEVAP EKGEELELSV ESPCELREPE PSPFSSKRTM YETEEMVIPG DPEEMRTFQS RTLALSRCSL EVILPSVHIF LPSKEVYESI YNRINNDLLM WEPADLLPTP DPAAQPSGFP GPSGFWHDSF KMCKSAFKLA NCFDLTPDSD SDDEDAHFFS VGASGGPQAA APEAPSLHLQ STFSTLVTVL KGRITALCET KDEGGKRLEA VHGELVLDME HGTLFSVSQY CGQPGLGYFC LEAEKATLYH RAAVDDYPLP SHLDLPSFAP PAQLAPTIYP SEEGVTERGA SGRKGQGRGP HMLSTAVRIH LDPHKNVKEF LVTLRLHKAT LRHYMALPEQ SWHSQLLEFL DVLDDPVLGY LPPTVITILH THLFSCSVDY RPLYLPVRVL ITAETFTLSS NIIMDTSTFL LRFILDDSAL YLSDKCEVET LDLRRDYVCV LDVDLLELVI KTWKGSTEGK LSQPLFELRC SNNVVHVHSC ADSCALLVNL LQYVMSTGDL HPPPRPPSPT EIAGQKLSES PASLPSCPPV ETALINQRDL ADALLDTERS LRELAQPSGG HLPQASPISV YLFPGERSGA PPPSPPVGGP AGSLGSCSEE KEDEREEEGD GDTLDSDEFC ILDAPGLGIP PRDGEPVVTQ LHPGPIVVRD GYFSRPIGST DLLRAPAHFP VPSTRVVLRE VSLVWHLYGG RDFGPHPGHR ARTGLSGPRS SPSRCSGPNR PQNSWRTQGG SGRQHHVLME IQLSKVSFQH EVYPAEPATG PAAPSQELEE RPLSRQVFIV QELEVRDRLA SSQINKFLYL HTSERMPRRA HSNMLTIKAL HVAPTTNLGG PECCLRVSLM PLRLNVDQDA LFFLKDFFTS LVAGINPVVP GETSAEARPE TRAQPSSPLE GQAEGVETTG SQEAPGGGHS PSPPDQQPIY FREFRFTSEV PIWLDYHGKH VTMDQVGTFA GLLIGLAQLN CSELKLKRLC CRHGLLGVDK VLGYALNEWL QDIRKNQLPG LLGGVGPMHS VVQLFQGFRD LLWLPIEQYR KDGRLMRGLQ RGAASFGSST ASAALELSNR LVQAIQATAE TVYDILSPAA PVSRSLQDKR SARRLRRGQQ PADLREGVAK AYDTVREGIL DTAQTICDVA SRGHEQKGLT GAVGGVIRQL PPTVVKPLIL ATEATSSLLG GMRNQIVPDA HKDHALKWRS DSAQDNSAVD GTAGPGSTGS R UniProtKB: Autophagy-related protein 2 homolog A |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM HEPES, 250 mM NaCl, 1 mM TCEP |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Details | Complex was reconstituted and subjected to size exclusion before grid prep. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: OTHER / Number grids imaged: 1 / Number real images: 1847 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)