-Search query
-Search result
Showing 1 - 50 of 72 items for (author: korkhov & b)
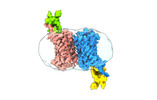
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
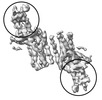
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)
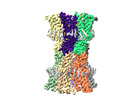
EMDB-18540: 
human connexin-36 gap junction channel in complex with mefloquine
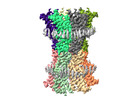
EMDB-18987: 
human connexin-36 gap junction channel
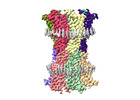
EMDB-18988: 
human connexin-36 gap junction channel in complex with quinine

PDB-8qoj: 
human connexin-36 gap junction channel in complex with mefloquine

PDB-8r7p: 
human connexin-36 gap junction channel

PDB-8r7q: 
human connexin-36 gap junction channel in complex with quinine

EMDB-17785: 
Cryo-EM structure of styrene oxide isomerase bound to benzylamine inhibitor

EMDB-17786: 
Cryo-EM structure of styrene oxide isomerase

PDB-8pnu: 
Cryo-EM structure of styrene oxide isomerase bound to benzylamine inhibitor

PDB-8pnv: 
Cryo-EM structure of styrene oxide isomerase

EMDB-16255: 
Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions

PDB-8bv5: 
Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions

EMDB-16249: 
Structure of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc

EMDB-16252: 
Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-16253: 
Focused refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-16254: 
Focused refinement of transmembrane domain of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

PDB-8buz: 
Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP

EMDB-15010: 
cryo-EM structure of Connexin 32 gap junction channel

EMDB-15011: 
cryo-EM structure of Connexin 32 gap junction channel

EMDB-15012: 
cryo-EM structure of Connexin 32 gap junction channel

EMDB-15013: 
cryo-EM structure of Connexin 32 R22G mutation gap junction channel

EMDB-15014: 
cryo-EM structure of Connexin 32 R22G mutation hemi channel

EMDB-15016: 
cryo-EM structure of Connexin 32 W3S mutation hemi channel

PDB-7zxm: 
cryo-EM structure of Connexin 32 gap junction channel

PDB-7zxn: 
cryo-EM structure of Connexin 32 gap junction channel

PDB-7zxo: 
cryo-EM structure of Connexin 32 gap junction channel

PDB-7zxp: 
cryo-EM structure of Connexin 32 R22G mutation gap junction channel

PDB-7zxq: 
cryo-EM structure of Connexin 32 R22G mutation hemi channel

PDB-7zxt: 
cryo-EM structure of Connexin 32 W3S mutation hemi channel
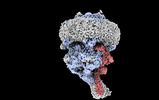
EMDB-16311: 
Structure of the rod CNG channel bound to calmodulin
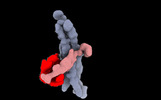
EMDB-16313: 
The structure of the rod CNG channel bound to calmodulin

PDB-8bx7: 
Structure of the rod CNG channel bound to calmodulin

EMDB-14452: 
Connexin43 gap junction channel structure in digitonin

EMDB-14455: 
Connexin43 gap junction channel structure in nanodisc

EMDB-14456: 
Connexin43 gap junction channel structure in digitonin

PDB-7z1t: 
Connexin43 gap junction channel structure in digitonin

PDB-7z22: 
Connexin43 gap junction channel structure in nanodisc

PDB-7z23: 
Connexin43 hemi channel in nanodisc

EMDB-14716: 
Structure of human OCT3 in lipid nanodisc
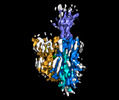
EMDB-14725: 
Structure of human OCT3 in complex with inhibitor Corticosterone
EMDB-14728: 
Structure of human OCT3 in complex with inhibitor decynium-22

PDB-7zh0: 
Structure of human OCT3 in lipid nanodisc

PDB-7zh6: 
Structure of human OCT3 in complex with inhibitor Corticosterone

PDB-7zha: 
Structure of human OCT3 in complex with inhibitor decynium-22

EMDB-14388: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya

PDB-7yzi: 
Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
