+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the rod CNG channel bound to calmodulin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
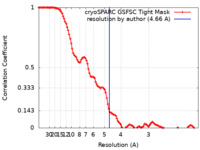
| Method | single particle reconstruction / cryo EM / Resolution: 4.66 Å | |||||||||
 Authors Authors | Marino J | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structural basis of calmodulin modulation of the rod cyclic nucleotide-gated channel. Authors: Diane C A Barret / Dina Schuster / Matthew J Rodrigues / Alexander Leitner / Paola Picotti / Gebhard F X Schertler / U Benjamin Kaupp / Volodymyr M Korkhov / Jacopo Marino /   Abstract: Calmodulin (CaM) regulates many ion channels to control calcium entry into cells, and mutations that alter this interaction are linked to fatal diseases. The structural basis of CaM regulation ...Calmodulin (CaM) regulates many ion channels to control calcium entry into cells, and mutations that alter this interaction are linked to fatal diseases. The structural basis of CaM regulation remains largely unexplored. In retinal photoreceptors, CaM binds to the CNGB subunit of cyclic nucleotide-gated (CNG) channels and, thereby, adjusts the channel's Cyclic guanosine monophosphate (cGMP) sensitivity in response to changes in ambient light conditions. Here, we provide the structural characterization for CaM regulation of a CNG channel by using a combination of single-particle cryo-electron microscopy and structural proteomics. CaM connects the CNGA and CNGB subunits, resulting in structural changes both in the cytosolic and transmembrane regions of the channel. Cross-linking and limited proteolysis-coupled mass spectrometry mapped the conformational changes induced by CaM in vitro and in the native membrane. We propose that CaM is a constitutive subunit of the rod channel to ensure high sensitivity in dim light. Our mass spectrometry-based approach is generally relevant for studying the effect of CaM on ion channels in tissues of medical interest, where only minute quantities are available. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16313.map.gz emd_16313.map.gz | 61.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16313-v30.xml emd-16313-v30.xml emd-16313.xml emd-16313.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16313_fsc.xml emd_16313_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16313.png emd_16313.png | 27.2 KB | ||
| Others |  emd_16313_half_map_1.map.gz emd_16313_half_map_1.map.gz emd_16313_half_map_2.map.gz emd_16313_half_map_2.map.gz | 116 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16313 http://ftp.pdbj.org/pub/emdb/structures/EMD-16313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16313 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16313.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16313.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||
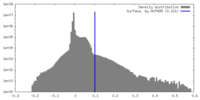
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16313_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
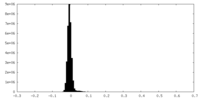
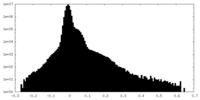
| Density Histograms |
-Half map: #2
| File | emd_16313_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
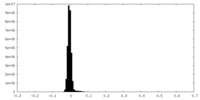
| Density Histograms |
- Sample components
Sample components
-Entire : Native CNG channel from retinal rods
| Entire | Name: Native CNG channel from retinal rods |
|---|---|
| Components |
|
-Supramolecule #1: Native CNG channel from retinal rods
| Supramolecule | Name: Native CNG channel from retinal rods / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 410 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.0 nm |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)