[English] 日本語
 Yorodumi
Yorodumi- EMDB-14388: Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Adenylyl cyclase / membrane protein / cyclic adenosine monophosphate / signal transduction / nanobody | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationreceptor guanylyl cyclase signaling pathway / peptide receptor activity / cGMP biosynthetic process / guanylate cyclase activity / adenylate cyclase / cAMP biosynthetic process / adenylate cyclase activity / manganese ion binding / intracellular signal transduction / magnesium ion binding ...receptor guanylyl cyclase signaling pathway / peptide receptor activity / cGMP biosynthetic process / guanylate cyclase activity / adenylate cyclase / cAMP biosynthetic process / adenylate cyclase activity / manganese ion binding / intracellular signal transduction / magnesium ion binding / ATP binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) / Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) /  | ||||||||||||
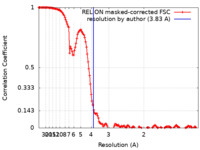
| Method | single particle reconstruction / cryo EM / Resolution: 3.83 Å | ||||||||||||
 Authors Authors | Mehta V / Khanppnavar B | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structure of Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases. Authors: Ved Mehta / Basavraj Khanppnavar / Dina Schuster / Ilayda Kantarci / Irene Vercellino / Angela Kosturanova / Tarun Iype / Sasa Stefanic / Paola Picotti / Volodymyr M Korkhov /  Abstract: adenylyl cyclase (AC) Rv1625c/Cya is an evolutionary ancestor of the mammalian membrane ACs and a model system for studies of their structure and function. Although the vital role of ACs in cellular ... adenylyl cyclase (AC) Rv1625c/Cya is an evolutionary ancestor of the mammalian membrane ACs and a model system for studies of their structure and function. Although the vital role of ACs in cellular signalling is well established, the function of their transmembrane (TM) regions remains unknown. Here, we describe the cryo-EM structure of Cya bound to a stabilizing nanobody at 3.6 Å resolution. The TM helices 1-5 form a structurally conserved domain that facilitates the assembly of the helical and catalytic domains. The TM region contains discrete pockets accessible from the extracellular and cytosolic side of the membrane. Neutralization of the negatively charged extracellular pocket Ex1 destabilizes the cytosolic helical domain and reduces the catalytic activity of the enzyme. The TM domain acts as a functional component of Cya, guiding the assembly of the catalytic domain and providing the means for direct regulation of catalytic activity in response to extracellular ligands. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14388.map.gz emd_14388.map.gz | 189.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14388-v30.xml emd-14388-v30.xml emd-14388.xml emd-14388.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14388_fsc.xml emd_14388_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14388.png emd_14388.png | 78.6 KB | ||
| Filedesc metadata |  emd-14388.cif.gz emd-14388.cif.gz | 6.4 KB | ||
| Others |  emd_14388_half_map_1.map.gz emd_14388_half_map_1.map.gz emd_14388_half_map_2.map.gz emd_14388_half_map_2.map.gz | 193.5 MB 193.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14388 http://ftp.pdbj.org/pub/emdb/structures/EMD-14388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14388 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14388 | HTTPS FTP |
-Related structure data
| Related structure data |  7yziMC  7yz9C  7yzkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14388.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14388.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
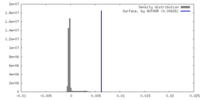
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14388_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
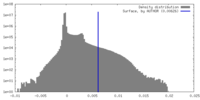
| Density Histograms |
-Half map: #1
| File | emd_14388_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
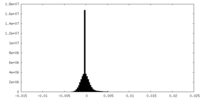
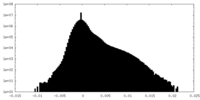
| Density Histograms |
- Sample components
Sample components
-Entire : A complex of Rv1625c / Cya homodimer bound to nano body Nb4
| Entire | Name: A complex of Rv1625c / Cya homodimer bound to nano body Nb4 |
|---|---|
| Components |
|
-Supramolecule #1: A complex of Rv1625c / Cya homodimer bound to nano body Nb4
| Supramolecule | Name: A complex of Rv1625c / Cya homodimer bound to nano body Nb4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Adenylate cyclase
| Supramolecule | Name: Adenylate cyclase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) |
-Supramolecule #3: Nanobody Nb4
| Supramolecule | Name: Nanobody Nb4 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Adenylate cyclase
| Macromolecule | Name: Adenylate cyclase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: adenylate cyclase |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) Mycobacterium tuberculosis '98-R604 INH-RIF-EM' (bacteria) |
| Molecular weight | Theoretical: 50.386613 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSAWSHPQF EKGSGLEVLF QGPSGHMAAR KCGAPPIAAD GSTRRPDCVT AVRTQARAPT QHYAESVARR QRVLTITAWL AVVVTGSFA LMQLATGAGG WYIALINVFT AVTFAIVPLL HRFGGLVAPL TFIGTAYVAI FAIGWDVGTD AGAQFFFLVA A ALVVLLVG ...String: MGSAWSHPQF EKGSGLEVLF QGPSGHMAAR KCGAPPIAAD GSTRRPDCVT AVRTQARAPT QHYAESVARR QRVLTITAWL AVVVTGSFA LMQLATGAGG WYIALINVFT AVTFAIVPLL HRFGGLVAPL TFIGTAYVAI FAIGWDVGTD AGAQFFFLVA A ALVVLLVG IEHTALAVGL AAVAAGLVIA LEFLVPPDTG LQPPWAMSVS FVLTTVSACG VAVATVWFAL RDTARAEAVM EA EHDRSEA LLANMLPASI AERLKEPERN IIADKYDEAS VLFADIVGFT ERASSTAPAD LVRFLDRLYS AFDELVDQHG LEK IKVSGD SYMVVSGVPR PRPDHTQALA DFALDMTNVA AQLKDPRGNP VPLRVGLATG PVVAGVVGSR RFFYDVWGDA VNVA SRMES TDSVGQIQVP DEVYERLKDD FVLRERGHIN VKGKGVMRTW YLIGRKVAAD PGEVRGAEPR TAGVAAA UniProtKB: Adenylate cyclase |
-Macromolecule #2: Nanobody Nb4
| Macromolecule | Name: Nanobody Nb4 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.771179 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQWQLVESG GGLVQAGGSL RLSCTASGII LSINSMGWYR QTAGNEREWV AFSTAGGSTT YADSVKGRFT ISRDNAKNTV YLQMNSLKP EDTAVYYCNT PAGRVGGTWG QGTPVTVSSH HHHHHEPEA |
-Macromolecule #3: 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: ONM |
|---|---|
| Molecular weight | Theoretical: 656.328 Da |
| Chemical component information | 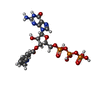 ChemComp-ONM: |
-Macromolecule #4: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM Tris pH 7.5, 200 mM NaCl, 0.1 % digitonin |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)