+Search query
-Structure paper
| Title | Structural basis of organic cation transporter-3 inhibition. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 6714, Year 2022 |
| Publish date | Nov 7, 2022 |
 Authors Authors | Basavraj Khanppnavar / Julian Maier / Freja Herborg / Ralph Gradisch / Erika Lazzarin / Dino Luethi / Jae-Won Yang / Chao Qi / Marion Holy / Kathrin Jäntsch / Oliver Kudlacek / Klaus Schicker / Thomas Werge / Ulrik Gether / Thomas Stockner / Volodymyr M Korkhov / Harald H Sitte /    |
| PubMed Abstract | Organic cation transporters (OCTs) facilitate the translocation of catecholamines, drugs and xenobiotics across the plasma membrane in various tissues throughout the human body. OCT3 plays a key role ...Organic cation transporters (OCTs) facilitate the translocation of catecholamines, drugs and xenobiotics across the plasma membrane in various tissues throughout the human body. OCT3 plays a key role in low-affinity, high-capacity uptake of monoamines in most tissues including heart, brain and liver. Its deregulation plays a role in diseases. Despite its importance, the structural basis of OCT3 function and its inhibition has remained enigmatic. Here we describe the cryo-EM structure of human OCT3 at 3.2 Å resolution. Structures of OCT3 bound to two inhibitors, corticosterone and decynium-22, define the ligand binding pocket and reveal common features of major facilitator transporter inhibitors. In addition, we relate the functional characteristics of an extensive collection of previously uncharacterized human genetic variants to structural features, thereby providing a basis for understanding the impact of OCT3 polymorphisms. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:36344565 / PubMed:36344565 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.2 - 3.67 Å |
| Structure data | EMDB-14716, PDB-7zh0: EMDB-14725, PDB-7zh6: EMDB-14728, PDB-7zha: |
| Chemicals | 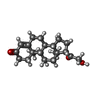 ChemComp-C0R: 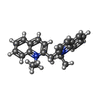 ChemComp-JDW: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Organic cation transporter / major-facilitator superfamily / SLC22 family / Corticosterone sensitive protein / membrane transport / Corticosterone-sensitive / Steroid hormones / Drug transporter / decynium-22 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









 homo sapiens (human)
homo sapiens (human)