+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human connexin-36 gap junction channel in complex with quinine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions / connexin complex / Gap junction assembly / gap junction channel activity / neuronal action potential / visual perception / cell-cell signaling / chemical synaptic transmission / synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.78 Å | |||||||||
 Authors Authors | Ding XY / Blum TB / Korkhov VM | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural basis of connexin-36 gap junction channel inhibition. Authors: Xinyue Ding / Simone Aureli / Anand Vaithia / Pia Lavriha / Dina Schuster / Basavraj Khanppnavar / Xiaodan Li / Thorsten B Blum / Paola Picotti / Francesco L Gervasio / Volodymyr M Korkhov /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18988.map.gz emd_18988.map.gz | 225.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18988-v30.xml emd-18988-v30.xml emd-18988.xml emd-18988.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18988.png emd_18988.png | 74.6 KB | ||
| Filedesc metadata |  emd-18988.cif.gz emd-18988.cif.gz | 5.3 KB | ||
| Others |  emd_18988_half_map_1.map.gz emd_18988_half_map_1.map.gz emd_18988_half_map_2.map.gz emd_18988_half_map_2.map.gz | 191.2 MB 190.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18988 http://ftp.pdbj.org/pub/emdb/structures/EMD-18988 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18988 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18988 | HTTPS FTP |
-Related structure data
| Related structure data |  8r7qMC  8qojC  8r7pC  8r7rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18988.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18988.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
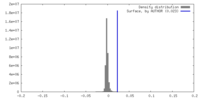
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18988_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
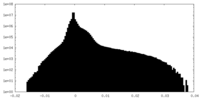
| Projections & Slices |
| ||||||||||||
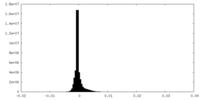
| Density Histograms |
-Half map: #1
| File | emd_18988_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
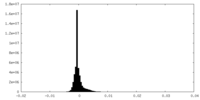
| Density Histograms |
- Sample components
Sample components
-Entire : human connexin-36 gap junction channel in complex with quinine
| Entire | Name: human connexin-36 gap junction channel in complex with quinine |
|---|---|
| Components |
|
-Supramolecule #1: human connexin-36 gap junction channel in complex with quinine
| Supramolecule | Name: human connexin-36 gap junction channel in complex with quinine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction delta-2 protein
| Macromolecule | Name: Gap junction delta-2 protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.078766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGEWTILERL LEAAVQQHST MIGRILLTVV VIFRILIVAI VGETVYDDEQ TMFVCNTLQP GCNQACYDRA FPISHIRYWV FQIIMVCTP SLCFITYSVH QSAKQRERRY STVFLALDRD PPESIGGPGG TGGGGSGGGK REDKKLQNAI VNGVLQNTEN T SKETEPDC ...String: MGEWTILERL LEAAVQQHST MIGRILLTVV VIFRILIVAI VGETVYDDEQ TMFVCNTLQP GCNQACYDRA FPISHIRYWV FQIIMVCTP SLCFITYSVH QSAKQRERRY STVFLALDRD PPESIGGPGG TGGGGSGGGK REDKKLQNAI VNGVLQNTEN T SKETEPDC LEVKELTPHP SGLRTASKSK LRRQEGISRF YIIQVVFRNA LEIGFLVGQY FLYGFSVPGL YECNRYPCIK EV ECYVSRP TEKTVFLVFM FAVSGICVVL NLAELNHLGW RKIKLAVRGA QAKRKSIYEI RNKDLPRVSV PNFGRTQSSD SAY VAAALE VLFQ UniProtKB: Gap junction delta-2 protein |
-Macromolecule #2: Quinine
| Macromolecule | Name: Quinine / type: ligand / ID: 2 / Number of copies: 12 / Formula: QI9 |
|---|---|
| Molecular weight | Theoretical: 324.417 Da |
| Chemical component information | 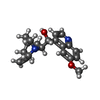 ChemComp-QI9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.78 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 231305 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)