-Search query
-Search result
Showing all 46 items for (author: koehl & a)

EMDB-18541: 
Structure of the mu opioid receptor bound to the antagonist nanobody NbE

PDB-8qot: 
Structure of the mu opioid receptor bound to the antagonist nanobody NbE

EMDB-41069: 
CDPPB-bound inactive mGlu5

EMDB-41092: 
Quis-bound intermediate mGlu5
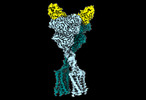
EMDB-41099: 
Quis-bound intermediate mGlu5
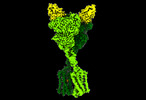
EMDB-41139: 
Quis and CDPPB bound active mGlu5

PDB-8t6j: 
CDPPB-bound inactive mGlu5

PDB-8t7h: 
Quis-bound intermediate mGlu5

PDB-8t8m: 
Quis-bound intermediate mGlu5

PDB-8tao: 
Quis and CDPPB bound active mGlu5

EMDB-13149: 
Mammalian orthoreovirus T3SA- in complex with the neural receptor NgR1

EMDB-13150: 
Mammalian orthoreovirus T3SA-

EMDB-25921: 
CryoET of presequence protease single particle

EMDB-22278: 
CryoEM structure of human presequence protease in partial open state 1

EMDB-22279: 
CryoEM structure of human presequence protease in partial open state 2

EMDB-22280: 
CryoEM structure of human presequence protease in open state

EMDB-22281: 
CryoEM structure of human presequence protease in partial closed state 1

PDB-6xos: 
CryoEM structure of human presequence protease in partial open state 1

PDB-6xot: 
CryoEM structure of human presequence protease in partial open state 2

PDB-6xou: 
CryoEM structure of human presequence protease in open state

PDB-6xov: 
CryoEM structure of human presequence protease in partial closed state 1

EMDB-11660: 
Structure of VP40 matrix layer in EBOV NP-VP24-VP35-VP40 VLPs

EMDB-11661: 
Structure of VP40 matrix layer in EBOV VP40 VLPs

EMDB-11662: 
Structure of VP40 matrix layer in EBOV VP40-GP VLPs

EMDB-11663: 
Structure of VP40 matrix layer in intact MARV virions

EMDB-11664: 
Structure of VP40 matrix layer in MARV VP40 VLPs

EMDB-11665: 
Structure of GP in EBOV VP40-GP VLPs

EMDB-0347: 
Apo form metabotropic glutamate receptor 5 with Nanobody 43

EMDB-0345: 
Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43

EMDB-0346: 
Metabotropic Glutamate Receptor 5 Apo Form

PDB-6n51: 
Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43

PDB-6n52: 
Metabotropic Glutamate Receptor 5 Apo Form

EMDB-7868: 
Mu Opioid Receptor-Gi Protein Complex

EMDB-7869: 
Mu Opioid Receptor-Gi Protein Complex

PDB-6dde: 
Mu Opioid Receptor-Gi Protein Complex

PDB-6ddf: 
Mu Opioid Receptor-Gi Protein Complex

EMDB-3870: 
Cryo-electron tomogram of Ebola virus nucleoprotein, residues 1-450

EMDB-3872: 
Cryo-electron tomogram of Ebola virus virus-like particles

EMDB-3873: 
The structure of Ebola virus nucleocapsid from virions

EMDB-3874: 
Cryo-electron tomogram of Ebola virus

EMDB-3875: 
The structure of Marburg virus nucleocapsid from virions

EMDB-3876: 
Cryo-electron tomogram of Marburg virus

EMDB-3869: 
The structure of Ebola virus nucleoprotein (residues 1-450) in nucleocapsid-like assemblies

EMDB-3871: 
The structure of Ebola virus nucleocapsid-like assemblies from recombinant virus-like particles (nucleoprotein, VP24,VP35,VP40)

PDB-6ehl: 
Model of the Ebola virus nucleoprotein in recombinant nucleocapsid-like assemblies

PDB-6ehm: 
Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
