+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0346 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Metabotropic Glutamate Receptor 5 Apo Form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cell Surface Receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / operant conditioning / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia ...sensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / operant conditioning / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia / positive regulation of long-term neuronal synaptic plasticity / desensitization of G protein-coupled receptor signaling pathway / positive regulation of sensory perception of pain / negative regulation of excitatory postsynaptic potential / positive regulation of dopamine secretion / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / positive regulation of neural precursor cell proliferation / glutamate receptor activity / nuclear inner membrane / Neurexins and neuroligins / astrocyte projection / response to corticosterone / response to morphine / temperature homeostasis / protein tyrosine kinase activator activity / conditioned place preference / regulation of synaptic transmission, glutamatergic / regulation of long-term synaptic depression / positive regulation of calcium-mediated signaling / protein tyrosine kinase binding / dendritic shaft / response to amphetamine / locomotory behavior / synapse organization / postsynaptic density membrane / G protein-coupled receptor activity / cognition / Schaffer collateral - CA1 synapse / cellular response to amyloid-beta / positive regulation of cytosolic calcium ion concentration / G alpha (q) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / learning or memory / positive regulation of MAPK cascade / response to antibiotic / neuronal cell body / dendrite / regulation of DNA-templated transcription / glutamatergic synapse / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Koehl A / Hu H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structural insights into the activation of metabotropic glutamate receptors. Authors: Antoine Koehl / Hongli Hu / Dan Feng / Bingfa Sun / Yan Zhang / Michael J Robertson / Matthew Chu / Tong Sun Kobilka / Toon Laeremans / Jan Steyaert / Jeffrey Tarrasch / Somnath Dutta / ...Authors: Antoine Koehl / Hongli Hu / Dan Feng / Bingfa Sun / Yan Zhang / Michael J Robertson / Matthew Chu / Tong Sun Kobilka / Toon Laeremans / Jan Steyaert / Jeffrey Tarrasch / Somnath Dutta / Rasmus Fonseca / William I Weis / Jesper M Mathiesen / Georgios Skiniotis / Brian K Kobilka /     Abstract: Metabotropic glutamate receptors are family C G-protein-coupled receptors. They form obligate dimers and possess extracellular ligand-binding Venus flytrap domains, which are linked by cysteine-rich ...Metabotropic glutamate receptors are family C G-protein-coupled receptors. They form obligate dimers and possess extracellular ligand-binding Venus flytrap domains, which are linked by cysteine-rich domains to their 7-transmembrane domains. Spectroscopic studies show that signalling is a dynamic process, in which large-scale conformational changes underlie the transmission of signals from the extracellular Venus flytraps to the G protein-coupling domains-the 7-transmembrane domains-in the membrane. Here, using a combination of X-ray crystallography, cryo-electron microscopy and signalling studies, we present a structural framework for the activation mechanism of metabotropic glutamate receptor subtype 5. Our results show that agonist binding at the Venus flytraps leads to a compaction of the intersubunit dimer interface, thereby bringing the cysteine-rich domains into close proximity. Interactions between the cysteine-rich domains and the second extracellular loops of the receptor enable the rigid-body repositioning of the 7-transmembrane domains, which come into contact with each other to initiate signalling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0346.map.gz emd_0346.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0346-v30.xml emd-0346-v30.xml emd-0346.xml emd-0346.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0346.png emd_0346.png | 146.8 KB | ||
| Filedesc metadata |  emd-0346.cif.gz emd-0346.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0346 http://ftp.pdbj.org/pub/emdb/structures/EMD-0346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0346 | HTTPS FTP |
-Related structure data
| Related structure data |  6n52MC  0345C  0347C  6n4xC  6n4yC  6n50C  6n51C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0346.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0346.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
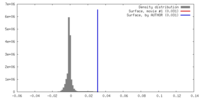
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Metabotropic Glutamate Receptor 5 in Nanodiscs
| Entire | Name: Metabotropic Glutamate Receptor 5 in Nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Metabotropic Glutamate Receptor 5 in Nanodiscs
| Supramolecule | Name: Metabotropic Glutamate Receptor 5 in Nanodiscs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: Metabotropic Glutamate Receptor 5 in Nanodiscs |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Metabotropic glutamate receptor 5
| Macromolecule | Name: Metabotropic glutamate receptor 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 97.677945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAAAQSSE RRVVAHMPGD IIIGALFSVH HQPTVDKVHE RKCGAVREQY GIQRVEAMLH TLERINSDP TLLPNITLGC EIRDSCWHSA VALEQSIEFI RDSLISSEEE EGLVRCVDGS SSSFRSKKPI VGVIGPGSSS V AIQVQNLL ...String: MKTIIALSYI FCLVFADYKD DDDAAAQSSE RRVVAHMPGD IIIGALFSVH HQPTVDKVHE RKCGAVREQY GIQRVEAMLH TLERINSDP TLLPNITLGC EIRDSCWHSA VALEQSIEFI RDSLISSEEE EGLVRCVDGS SSSFRSKKPI VGVIGPGSSS V AIQVQNLL QLFNIPQIAY SATSMDLSDK TLFKYFMRVV PSDAQQARAM VDIVKRYNWT YVSAVHTEGN YGESGMEAFK DM SAKEGIC IAHSYKIYSN AGEQSFDKLL KKLTSHLPKA RVVACFCEGM TVRGLLMAMR RLGLAGEFLL LGSDGWADRY DVT DGYQRE AVGGITIKLQ SPDVKWFDDY YLKLRPETNH RNPWFQEFWQ HRFQCRLEGF PQENSKYNKT CNSSLTLKTH HVQD SKMGF VINAIYSMAY GLHNMQMSLC PGYAGLCDAM KPIDGRKLLE SLMKTNFTGV SGDTILFDEN GDSPGRYEIM NFKEM GKDY FDYINVGSWD NGELKMDDDE VWSKKSNIIR SVCSEPCEKG QIKVIRKGEV SCCWTCTPCK ENEYVFDEYT CKACQL GSW PTDDLTGCDL IPVQYLRWGD PEPIAAVVFA CLGLLATLFV TVVFIIYRDT PVVKSSSREL CYIILAGICL GYLCTFC LI AKPKQIYCYL QRIGIGLSPA MSYSALVTKT NRIARILAGS KKKICTKKPR FMSACAQLVI AFILICIQLG IIVALFIM E PPDIMHDYPS IREVYLICNT TNLGVVTPLG YNGLLILSCT FYAFKTRNVP ANFNEAKYIA FTMYTTCIIW LAFVPIYFG SNYKIITMCF SVSLSATVAL GCMFVPKVYI ILAKPERNVR SAFTTSTVVR MHVGDGKSSS AASRSSSLVN L UniProtKB: Metabotropic glutamate receptor 5 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 100mM NaCl, 20 mM HEPES pH 7.5, 5uM FFMTEB added as a negative allosteric modulator. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: Addition 0.0005% Amphipol A8-35was added to sample prior to apply to grid; 3.5ul sample was applied; blot for 1s before plunging. | |||||||||
| Details | This sample was mono disperse as assayed by gel filtration. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 47169 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab initio model was generated from VIPER. |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 123096 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 2.1) |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)