+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3873 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of Ebola virus nucleocapsid from virions | ||||||||||||
 Map data Map data | Subtomogram averaging of Ebola virus nucleocapsid within intact virions | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 9.1 Å | ||||||||||||
 Authors Authors | Wan W / Kolesnikova L / Clarke M / Koehler A / Noda T / Becker S / Briggs JAG | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure and assembly of the Ebola virus nucleocapsid. Authors: William Wan / Larissa Kolesnikova / Mairi Clarke / Alexander Koehler / Takeshi Noda / Stephan Becker / John A G Briggs /    Abstract: Ebola and Marburg viruses are filoviruses: filamentous, enveloped viruses that cause haemorrhagic fever. Filoviruses are within the order Mononegavirales, which also includes rabies virus, measles ...Ebola and Marburg viruses are filoviruses: filamentous, enveloped viruses that cause haemorrhagic fever. Filoviruses are within the order Mononegavirales, which also includes rabies virus, measles virus, and respiratory syncytial virus. Mononegaviruses have non-segmented, single-stranded negative-sense RNA genomes that are encapsidated by nucleoprotein and other viral proteins to form a helical nucleocapsid. The nucleocapsid acts as a scaffold for virus assembly and as a template for genome transcription and replication. Insights into nucleoprotein-nucleoprotein interactions have been derived from structural studies of oligomerized, RNA-encapsidating nucleoprotein, and cryo-electron microscopy of nucleocapsid or nucleocapsid-like structures. There have been no high-resolution reconstructions of complete mononegavirus nucleocapsids. Here we apply cryo-electron tomography and subtomogram averaging to determine the structure of Ebola virus nucleocapsid within intact viruses and recombinant nucleocapsid-like assemblies. These structures reveal the identity and arrangement of the nucleocapsid components, and suggest that the formation of an extended α-helix from the disordered carboxy-terminal region of nucleoprotein-core links nucleoprotein oligomerization, nucleocapsid condensation, RNA encapsidation, and accessory protein recruitment. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3873.map.gz emd_3873.map.gz | 24.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3873-v30.xml emd-3873-v30.xml emd-3873.xml emd-3873.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3873.png emd_3873.png | 149.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3873 http://ftp.pdbj.org/pub/emdb/structures/EMD-3873 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3873 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3873 | HTTPS FTP |
-Related structure data
| Related structure data |  3869C  3870C  3871C  3872C  3874C  3875C  3876C  6ehlC  6ehmC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3873.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3873.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subtomogram averaging of Ebola virus nucleocapsid within intact virions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
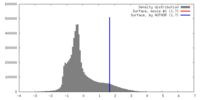
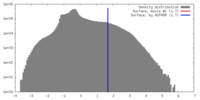
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ebola virus - Mayinga, Zaire, 1976
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Ebola virus - Mayinga, Zaire, 1976
| Supramolecule | Name: Ebola virus - Mayinga, Zaire, 1976 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Virus was isolated from infected VeroE6 cells. Purified viruses were fixed with paraformaldehyde. NCBI-ID: 128952 / Sci species name: Ebola virus - Mayinga, Zaire, 1976 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Nucleocapsid / Diameter: 280.0 Å |
-Macromolecule #1: Ebola virus nucleoprotein
| Macromolecule | Name: Ebola virus nucleoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MDSRPQKIWM APSLTESDMD YHKILTAGLS VQQGIVRQRV IPVYQVNNLE EICQLIIQAF EAGVDFQES ADSFLLMLCL HHAYQGDYKL FLESGAVKYL EGHGFRFEVK KRDGVKRLEE L LPAVSSGK NIKRTLAAMP EEETTEANAG QFLSFASLFL PKLVVGEKAC ...String: MDSRPQKIWM APSLTESDMD YHKILTAGLS VQQGIVRQRV IPVYQVNNLE EICQLIIQAF EAGVDFQES ADSFLLMLCL HHAYQGDYKL FLESGAVKYL EGHGFRFEVK KRDGVKRLEE L LPAVSSGK NIKRTLAAMP EEETTEANAG QFLSFASLFL PKLVVGEKAC LEKVQRQIQV HA EQGLIQY PTAWQSVGHM MVIFRLMRTN FLIKFLLIHQ GMHMVAGHDA NDAVISNSVA QAR FSGLLI VKTVLDHILQ KTERGVRLHP LARTAKVKNE VNSFKAALSS LAKHGEYAPF ARLL NLSGV NNLEHGLFPQ LSAIALGVAT AHGSTLAGVN VGEQYQQLRE AATEAEKQLQ QYAES RELD HLGLDDQEKK ILMNFHQKKN EISFQQTNAM VTLRKERLAK LTEAITAASL PKTSGH YDD DDDIPFPGPI NDDDNPGHQD DDPTDSQDTT IPDVVVDPDD GSYGEYQSYS ENGMNAP DD LVLFDLDEDD EDTKPVPNRS TKGGQQKNSQ KGQHIEGRQT QSRPIQNVPG PHRTIHHA S APLTDNDRRN EPSGSTSPRM LTPINEEADP LDDADDETSS LPPLESDDEE QDRDGTSNR TPTVAPPAPV YRDHSEKKEL PQDEQQDQDH TQEARNQDSD NTQSEHSFEE MYRHILRSQG PFDAVLYYH MMKDEPVVFS TSDGKEYTYP DSLEEEYPPW LTEKEAMNEE NRFVTLDGQQ F YWPVMNHK NKFMAILQHH Q |
-Macromolecule #2: Ebola virus VP24
| Macromolecule | Name: Ebola virus VP24 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MAKATGRYNL ISPKKDLEKG VVLSDLCNFL VSQTIQGWKV YWAGIEFDVT HKGMALLHRL KTNDFAPAW SMTRNLFPHL FQNPNSTIES PLWALRVILA AGIQDQLIDQ SLIEPLAGAL G LISDWLLT TNTNHFNMRT QRVKEQLSLK MLSLIRSNIL KFINKLDALH ...String: MAKATGRYNL ISPKKDLEKG VVLSDLCNFL VSQTIQGWKV YWAGIEFDVT HKGMALLHRL KTNDFAPAW SMTRNLFPHL FQNPNSTIES PLWALRVILA AGIQDQLIDQ SLIEPLAGAL G LISDWLLT TNTNHFNMRT QRVKEQLSLK MLSLIRSNIL KFINKLDALH VVNYNGLLSS IE IGTQNHT IIITRTNMGF LVELQEPDKS AMNRMKPGPA KFSLLHESTL KAFTQGSSTR MQS LILEFN SSLAI |
-Macromolecule #3: Ebola virus VP35
| Macromolecule | Name: Ebola virus VP35 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MTTRTKGRGH TAATTQNDRM PGPELSGWIS EQLMTGRIPV SDIFCDIENN PGLCYASQMQ QTKPNPKTR NSQTQTDPIC NHSFEEVVQT LASLATVVQQ QTIASESLEQ RITSLENGLK P VYDMAKTI SSLNRVCAEM VAKYDLLVMT TGRATATAAA TEAYWAEHGQ ...String: MTTRTKGRGH TAATTQNDRM PGPELSGWIS EQLMTGRIPV SDIFCDIENN PGLCYASQMQ QTKPNPKTR NSQTQTDPIC NHSFEEVVQT LASLATVVQQ QTIASESLEQ RITSLENGLK P VYDMAKTI SSLNRVCAEM VAKYDLLVMT TGRATATAAA TEAYWAEHGQ PPPGPSLYEE SA IRGKIES RDETVPQSVR EAFNNLNSTT SLTEENFGKP DISAKDLRNI MYDHLPGFGT AFH QLVQVI CKLGKDSNSL DIIHAEFQAS LAEGDSPQCA LIQITKRVPI FQDAAPPVIH IRSR GDIPR ACQKSLRPVP PSPKIDRGWV CVFQLQDGKT LGLKI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: Virus was purified into Dulbecco's modified Eagle's medium (DMEM) with 4% paraformaldehyde |
|---|---|
| Grid | Model: C-flat 2/1 3C / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3708 pixel / Digitization - Dimensions - Height: 3708 pixel / Digitization - Frames/image: 1-5 / Average exposure time: 2.0 sec. / Average electron dose: 2.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)