+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CDPPB-bound inactive mGlu5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / operant conditioning / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia ...sensory perception of hot stimulus / A2A adenosine receptor binding / : / negative regulation of dendritic spine morphogenesis / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / operant conditioning / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / protein localization to nuclear inner membrane / positive regulation of cellular response to hypoxia / positive regulation of long-term neuronal synaptic plasticity / desensitization of G protein-coupled receptor signaling pathway / positive regulation of sensory perception of pain / negative regulation of excitatory postsynaptic potential / positive regulation of dopamine secretion / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / positive regulation of neural precursor cell proliferation / glutamate receptor activity / nuclear inner membrane / Neurexins and neuroligins / astrocyte projection / response to corticosterone / response to morphine / temperature homeostasis / protein tyrosine kinase activator activity / conditioned place preference / regulation of synaptic transmission, glutamatergic / regulation of long-term synaptic depression / positive regulation of calcium-mediated signaling / protein tyrosine kinase binding / dendritic shaft / response to amphetamine / locomotory behavior / synapse organization / postsynaptic density membrane / G protein-coupled receptor activity / cognition / Schaffer collateral - CA1 synapse / cellular response to amyloid-beta / positive regulation of cytosolic calcium ion concentration / G alpha (q) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / learning or memory / positive regulation of MAPK cascade / response to antibiotic / neuronal cell body / dendrite / regulation of DNA-templated transcription / glutamatergic synapse / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Krishna Kumar K / Wang H / Kobilka BK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2023 Title: Step-wise activation of a Family C GPCR. Authors: Kaavya Krishna Kumar / Haoqing Wang / Chris Habrian / Naomi R Latorraca / Jun Xu / Evan S O'Brien / Chensong Zhang / Elizabeth Montabana / Antoine Koehl / Susan Marqusee / Ehud Y Isacoff / Brian K Kobilka /  Abstract: Metabotropic glutamate receptors belong to a family of G protein-coupled receptors that are obligate dimers and possess a large extracellular ligand-binding domain (ECD) that is linked via a cysteine- ...Metabotropic glutamate receptors belong to a family of G protein-coupled receptors that are obligate dimers and possess a large extracellular ligand-binding domain (ECD) that is linked via a cysteine-rich domain (CRDs) to their 7-transmembrane (TM) domain. Upon activation, these receptors undergo a large conformational change to transmit the ligand binding signal from the ECD to the G protein-coupling TM. In this manuscript, we propose a model for a sequential, multistep activation mechanism of metabotropic glutamate receptor subtype 5. We present a series of structures in lipid nanodiscs, from inactive to fully active, including agonist-bound intermediate states. Further, using bulk and single-molecule fluorescence imaging we reveal distinct receptor conformations upon allosteric modulator and G protein binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41069.map.gz emd_41069.map.gz | 114.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41069-v30.xml emd-41069-v30.xml emd-41069.xml emd-41069.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41069.png emd_41069.png | 41 KB | ||
| Filedesc metadata |  emd-41069.cif.gz emd-41069.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41069 http://ftp.pdbj.org/pub/emdb/structures/EMD-41069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41069 | HTTPS FTP |
-Related structure data
| Related structure data |  8t6jMC  8t7hC  8t8mC  8taoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41069.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41069.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8521 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Metabotropic glutamate receptor 5 in complex with CDPPB
| Entire | Name: Metabotropic glutamate receptor 5 in complex with CDPPB |
|---|---|
| Components |
|
-Supramolecule #1: Metabotropic glutamate receptor 5 in complex with CDPPB
| Supramolecule | Name: Metabotropic glutamate receptor 5 in complex with CDPPB type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Metabotropic glutamate receptor 5
| Macromolecule | Name: Metabotropic glutamate receptor 5 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.868297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAAQSSER RVVAHMPGDI IIGALFSVHH QPTVDKVHER KCGAVREQYG IQRVEAMLHT LERINSDPT LLPNITLGCE IRDSCWHSAV ALEQSIEFIR DSLISSEEEE GLVRCVDGSS SSFRSKKPIV GVIGPGSSSV A IQVQNLLQ ...String: MKTIIALSYI FCLVFADYKD DDDAAQSSER RVVAHMPGDI IIGALFSVHH QPTVDKVHER KCGAVREQYG IQRVEAMLHT LERINSDPT LLPNITLGCE IRDSCWHSAV ALEQSIEFIR DSLISSEEEE GLVRCVDGSS SSFRSKKPIV GVIGPGSSSV A IQVQNLLQ LFNIPQIAYS ATSMDLSDKT LFKYFMRVVP SDAQQARAMV DIVKRYNWTY VSAVHTEGNY GESGMEAFKD MS AKEGICI AHSYKIYSNA GEQSFDKLLK KLTSHLPKAR VVACFCEGMT VRGLLMAMRR LGLAGEFLLL GSDGWADRYD VTD GYQREA VGGITIKLQS PDVKWFDDYY LKLRPETNHR NPWFQEFWQH RFQCRLEGFP QENSKYNKTC NSSLTLKTHH VQDS KMGFV INAIYSMAYG LHNMQMSLCP GYAGLCDAMK PIDGRKLLES LMKTNFTGVS GDTILFDENG DSPGRYEIMN FKEMG KDYF DYINVGSWDN GELKMDDDEV WSKKSNIIRS VCSEPCEKGQ IKVIRKGEVS CCWTCTPCKE NEYVFDEYTC KACQLG SWP TDDLTGCDLI PVQYLRWGDP EPIAAVVFAC LGLLATLFVT VVFIIYRDTP VVKSSSRELC YIILAGICLG YLCTFCL IA KPKQIYCYLQ RIGIGLSPAM SYSALVTKTN RIARILAGSK KKICTKKPRF MSACAQLVIA FILICIQLGI IVALFIME P PDIMHDYPSI REVYLICNTT NLGVVTPLGY NGLLILSCTF YAFKTRNVPA NFNEAKYIAF TMYTTCIIWL AFVPIYFGS NYKIITMCFS VSLSATVALG CMFVPKVYII LAKPERNVRS AFTTSTVVRM HVGDGKSSSA ASRSSSLVNL WKRRGSSGET L UniProtKB: Metabotropic glutamate receptor 5 |
-Macromolecule #2: 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide
| Macromolecule | Name: 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide / type: ligand / ID: 2 / Number of copies: 2 / Formula: YKU |
|---|---|
| Molecular weight | Theoretical: 364.399 Da |
| Chemical component information | 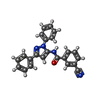 ChemComp-YKU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 246232 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















