-Search query
-Search result
Showing 1 - 50 of 62 items for (author: kim & gj)

EMDB-37593: 
Vibrio vulnificus MARTX effector duet (RDTND-RID) complexed with human Rac1 Q61L and calmodulin

EMDB-35904: 
AtSLAC1 8D mutant in closed state

EMDB-35920: 
AtSLAC1 in open state

PDB-8j0j: 
AtSLAC1 8D mutant in closed state

PDB-8j1e: 
AtSLAC1 in open state

EMDB-34303: 
AtSLAC1 6D mutant in closed state

EMDB-34304: 
AtSLAC1 6D mutant in open state

PDB-8gw6: 
AtSLAC1 6D mutant in closed state

PDB-8gw7: 
AtSLAC1 6D mutant in open state

EMDB-28728: 
Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

EMDB-28729: 
Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

EMDB-28730: 
Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

PDB-8ez3: 
Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

PDB-8ez7: 
Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

PDB-8ez8: 
Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
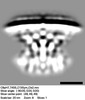
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
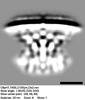
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask

EMDB-34530: 
Membrane protein A

EMDB-34531: 
Membrane protein B

EMDB-35713: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc

PDB-8h86: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc

PDB-8h87: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc

PDB-8iu0: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc

EMDB-34350: 
Legionella pneumophila Deubiquitinase, LotA(7-544) apo state

EMDB-25804: 
Cryo-EM structure of methane monooxygenase hydroxylase (by quantifoil)

EMDB-25805: 
Cryo-EM structure of methane monooxygenase hydroxylase (by graphene)

PDB-7tc7: 
Cryo-EM structure of methane monooxygenase hydroxylase (by quantifoil)

PDB-7tc8: 
Cryo-EM structure of methane monooxygenase hydroxylase (by graphene)

EMDB-24822: 
20S proteasome from red blood cell lysate

EMDB-26216: 
Methylococcus capsulatus methane monooxygenase hydroxylase (MMOH) using the plasma-jet treated grid

EMDB-10793: 
Cryo-EM structure of the Full-length disease type human Huntingtin

PDB-6yej: 
Cryo-EM structure of the Full-length disease type human Huntingtin

EMDB-30323: 
Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF

EMDB-30324: 
Cryo-EM structure of human GABA(B) receptor in apo state

EMDB-30472: 
Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626

PDB-7ca3: 
Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF

PDB-7ca5: 
Cryo-EM structure of human GABA(B) receptor in apo state

PDB-7cum: 
Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626

EMDB-21685: 
Structure of human GABA(B) receptor in an inactive state

PDB-6wiv: 
Structure of human GABA(B) receptor in an inactive state

EMDB-30220: 
AdhE spirosome in extended conformation

EMDB-4937: 
Cryo-EM 3D map of normal Huntingtin

EMDB-4944: 
Cryo-EM 3D map of the N-terminal GFP tagged normal type Huntingtin

PDB-6rmh: 
The Rigid-body refined model of the normal Huntingtin.

EMDB-0896: 
High resolution structure of FANCA C-terminal domain (CTD)

EMDB-0899: 
High resolution structure of FANCA C-terminal domain (CTD)

EMDB-0900: 
Structure of FANCA and FANCG Complex

EMDB-0901: 
Structure of N-terminal and C-terminal domains of FANCA

PDB-6lhs: 
High resolution structure of FANCA C-terminal domain (CTD)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
