[English] 日本語
 Yorodumi
Yorodumi- EMDB-28729: Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) inf... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | |||||||||||||||
 Map data Map data | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | antibody / neuraminidase / influenza / Hydrolase-Immune System complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationexo-alpha-sialidase / exo-alpha-sialidase activity / viral budding from plasma membrane / carbohydrate metabolic process / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) / Homo sapiens (human) /  Influenza A virus (A/Moscow/10/1999(H3N2)) Influenza A virus (A/Moscow/10/1999(H3N2)) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||||||||
 Authors Authors | Mou Z / Lei R / Wu NC / Dai X | |||||||||||||||
| Funding support | 4 items
| |||||||||||||||
 Citation Citation |  Journal: Immunity / Year: 2023 Journal: Immunity / Year: 2023Title: Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase. Authors: Ruipeng Lei / Wooseob Kim / Huibin Lv / Zongjun Mou / Michael J Scherm / Aaron J Schmitz / Jackson S Turner / Timothy J C Tan / Yiquan Wang / Wenhao O Ouyang / Weiwen Liang / Joel Rivera- ...Authors: Ruipeng Lei / Wooseob Kim / Huibin Lv / Zongjun Mou / Michael J Scherm / Aaron J Schmitz / Jackson S Turner / Timothy J C Tan / Yiquan Wang / Wenhao O Ouyang / Weiwen Liang / Joel Rivera-Cardona / Chuyun Teo / Claire S Graham / Christopher B Brooke / Rachel M Presti / Chris K P Mok / Florian Krammer / Xinghong Dai / Ali H Ellebedy / Nicholas C Wu /    Abstract: There is growing appreciation for neuraminidase (NA) as an influenza vaccine target; however, its antigenicity remains poorly characterized. In this study, we isolated three broadly reactive N2 ...There is growing appreciation for neuraminidase (NA) as an influenza vaccine target; however, its antigenicity remains poorly characterized. In this study, we isolated three broadly reactive N2 antibodies from the plasmablasts of a single vaccinee, including one that cross-reacts with NAs from seasonal H3N2 strains spanning five decades. Although these three antibodies have diverse germline usages, they recognize similar epitopes that are distant from the NA active site and instead involve the highly conserved underside of NA head domain. We also showed that all three antibodies confer prophylactic and therapeutic protection in vivo, due to both Fc effector functions and NA inhibition through steric hindrance. Additionally, the contribution of Fc effector functions to protection in vivo inversely correlates with viral growth inhibition activity in vitro. Overall, our findings advance the understanding of NA antibody response and provide important insights into the development of a broadly protective influenza vaccine. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28729.map.gz emd_28729.map.gz | 167.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28729-v30.xml emd-28729-v30.xml emd-28729.xml emd-28729.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28729.png emd_28729.png | 43.5 KB | ||
| Filedesc metadata |  emd-28729.cif.gz emd-28729.cif.gz | 7 KB | ||
| Others |  emd_28729_half_map_1.map.gz emd_28729_half_map_1.map.gz emd_28729_half_map_2.map.gz emd_28729_half_map_2.map.gz | 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28729 http://ftp.pdbj.org/pub/emdb/structures/EMD-28729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28729 | HTTPS FTP |
-Related structure data
| Related structure data |  8ez7MC  8ez3C  8ez8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28729.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28729.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
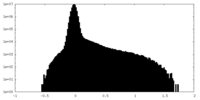
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_28729_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
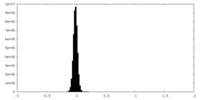
| Density Histograms |
-Half map: Half Map 2
| File | emd_28729_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
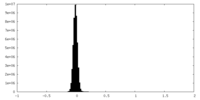
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) inf...
| Entire | Name: Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase |
|---|---|
| Components |
|
-Supramolecule #1: Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) inf...
| Supramolecule | Name: Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Macromolecule #1: Heavy chain of influenza virus neuraminidase antibody 1F04
| Macromolecule | Name: Heavy chain of influenza virus neuraminidase antibody 1F04 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.984465 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVLPGRSLRL SCVASGFDFS SFEMNWVRQA PGTGLEWLSY ISGNGDTIYY AESVKGRFTI SRDNAKNSLY LQMNGLRAE DTATYYCARQ RSVYSTSRSN WSVFLDDWGQ GTLVTVS |
-Macromolecule #2: Light chain of influenza virus neuraminidase antibody 1F04
| Macromolecule | Name: Light chain of influenza virus neuraminidase antibody 1F04 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.8201 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGESAT LSCRASQSLA PNYLAWFQQQ PGQAPRLLIY GASSRAAGIP DRFSGSGSGT DFTLTITRLE PEDFAVYYC QQYGGSPPYT FGQGTKVEIK R |
-Macromolecule #3: Neuraminidase
| Macromolecule | Name: Neuraminidase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: exo-alpha-sialidase |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Moscow/10/1999(H3N2)) Influenza A virus (A/Moscow/10/1999(H3N2)) |
| Molecular weight | Theoretical: 42.767715 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: AEYRNWSKPQ CNITGFAPFS KDNSIRLSAG GDIWVTREPY VSCDPDKCYQ FALGQGTTLN NGHSNDTVHD RTPYRTLLMN ELGVPFHLG TKQVCIAWSS SSCHDGKAWL HVCVTGDDEN ATASFIYNGR LVDSIGSWSK KILRTQESEC VCINGTCTVV M TDGSASGK ...String: AEYRNWSKPQ CNITGFAPFS KDNSIRLSAG GDIWVTREPY VSCDPDKCYQ FALGQGTTLN NGHSNDTVHD RTPYRTLLMN ELGVPFHLG TKQVCIAWSS SSCHDGKAWL HVCVTGDDEN ATASFIYNGR LVDSIGSWSK KILRTQESEC VCINGTCTVV M TDGSASGK ADTKILFIEE GKIVHTSPLS GSAQHVEECS CYPRYPGVRC VCRDNWKGSN RPIVDINVKD YSIVSSYVCS GL VGDTPRK NDSSSSSHCL DPNNEEGGHG VKGWAFDDGN DVWMGRTISE KLRSGYETFK VIEGWSKPNS KLQINRQVIV DRG NRSGYS GIFSVEGKSC INRCFYVELI RGRKQETEVL WTSNSIVVFC GTSGTYGTGS WPDGADINLM PI UniProtKB: Neuraminidase |
-Macromolecule #5: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 1 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)