[English] 日本語
 Yorodumi
Yorodumi- EMDB-30472: Cryo-EM structure of human GABA(B) receptor bound to the antagoni... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30472 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / GABA / Neurosignalling / Signaling protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion ...negative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / negative regulation of synaptic transmission / positive regulation of glutamate secretion / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / axolemma / dendritic shaft / response to nicotine / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / mitochondrial membrane / GABA-ergic synapse / Schaffer collateral - CA1 synapse / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / synaptic vesicle / osteoblast differentiation / transmembrane signaling receptor activity / presynaptic membrane / G alpha (i) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / postsynaptic membrane / neuron projection / G protein-coupled receptor signaling pathway / protein heterodimerization activity / negative regulation of cell population proliferation / neuronal cell body / endoplasmic reticulum membrane / glutamatergic synapse / extracellular space / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.52 Å | |||||||||
 Authors Authors | Kim Y / Jeong E | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Structural Basis for Activation of the Heterodimeric GABA Receptor. Authors: Yoojoong Kim / Eunyoung Jeong / Ji-Hong Jeong / Youngjin Kim / Yunje Cho /  Abstract: The neurotransmitter γ-aminobutyric acid (GABA) activates the metabotropic GABA receptor to generate slow, prolonged inhibitory signals that regulate the neural circuitry. The GABA receptor is an ...The neurotransmitter γ-aminobutyric acid (GABA) activates the metabotropic GABA receptor to generate slow, prolonged inhibitory signals that regulate the neural circuitry. The GABA receptor is an obligate heterodimeric G protein-coupled receptor (GPCR) comprised of GBR1 and GBR2 subunits, each with extracellular, seven-helix transmembrane (7TM), and coiled-coil domains. To understand how GABA-driven conformational changes in the extracellular domain are transmitted to the 7TM domain during signal transduction, we determined cryo-electron microscopy (EM) structures of GABA in two different states: an antagonist-bound inactive state, and an active state in which both the GABA agonist and a positive allosteric modulator (PAM) are bound. In the inactive state, the TM3 and TM5 helices in the two 7TM domains engage in cholesterol-mediated as well as direct interactions, resulting in an open conformation. GABA binding forces the extracellular domains of GBR1 and GBR2 into a compact form, relocating the linkers that connect the extracellular and 7TM domains closer to each other. The movement of the linker along with the associated extracellular loop 2 of the 7TM domain reorients the two 7TM domains and creates a new interface with the TM5, TM6 and TM7 helices in a closed conformation. PAM binding to the interface between the TM6 and TM6 helices stabilizes the active 7TM domain conformation. The relayed structural rearrangement results in significant conformational changes in the TM helices, as well as intracellular loop 3 in GBR2, which may promote the binding and activation of the Gi/o proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30472.map.gz emd_30472.map.gz | 42.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30472-v30.xml emd-30472-v30.xml emd-30472.xml emd-30472.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30472.png emd_30472.png | 55.3 KB | ||
| Filedesc metadata |  emd-30472.cif.gz emd-30472.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30472 http://ftp.pdbj.org/pub/emdb/structures/EMD-30472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30472 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30472 | HTTPS FTP |
-Related structure data
| Related structure data |  7cumMC  7ca3C  7ca5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30472.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30472.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Gamma-aminobutyric acid type B receptor
| Entire | Name: Gamma-aminobutyric acid type B receptor |
|---|---|
| Components |
|
-Supramolecule #1: Gamma-aminobutyric acid type B receptor
| Supramolecule | Name: Gamma-aminobutyric acid type B receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Antagonist-bound Gamma-aminobutyric acid type B receptor |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 200 kDa/nm |
-Macromolecule #1: Gamma-aminobutyric acid type B receptor subunit 1
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 87.248195 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRLLTALFAY FIVALILAFS VSAKSMSERR AVYIGALFPM SGGWPGGQAC QPAVEMALED VNSRRDILPD YELKLIHHDS KCDPGQATK YLYELLYNDP IKIILMPGCS SVSTLVAEAA RMWNLIVLSY GSSSPALSNR QRFPTFFRTH PSATLHNPTR V KLFEKWGW ...String: MRLLTALFAY FIVALILAFS VSAKSMSERR AVYIGALFPM SGGWPGGQAC QPAVEMALED VNSRRDILPD YELKLIHHDS KCDPGQATK YLYELLYNDP IKIILMPGCS SVSTLVAEAA RMWNLIVLSY GSSSPALSNR QRFPTFFRTH PSATLHNPTR V KLFEKWGW KKIATIQQTT EVFTSTLDDL EERVKEAGIE ITFRQSFFSD PAVPVKNLKR QDARIIVGLF YETEARKVFC EV YKERLFG KKYVWFLIGW YADNWFKIYD PSINCTVDEM TEAVEGHITT EIVMLNPANT RSISNMTSQE FVEKLTKRLK RHP EETGGF QEAPLAYDAI WALALALNKT SGGGGRSGVR LEDFNYNNQT ITDQIYRAMN SSSFEGVSGH VVFDASGSRM AWTL IEQLQ GGSYKKIGYY DSTKDDLSWS KTDKWIGGSP PADQTLVIKT FRFLSQKLFI SVSVLSSLGI VLAVVCLSFN IYNSH VRYI QNSQPNLNNL TAVGCSLALA AVFPLGLDGY HIGRNQFPFV CQARLWLLGL GFSLGYGSMF TKIWWVHTVF TKKEEK KEW RKTLEPWKLY ATVGLLVGMD VLTLAIWQIV DPLHRTIETF AKEEPKEDID VSILPQLEHC SSRKMNTWLG IFYGYKG LL LLLGIFLAYE TKSVSTEKIN DHRAVGMAIY NVAVLCLITA PVTMILSSQQ DAAFAFASLA IVFSSYITLV VLFVPKMR R LITRGEWQSE AQDTMKTGSS TNNNEEEKSR LLEKENRELE KSGRLEVLFQ UniProtKB: Gamma-aminobutyric acid type B receptor subunit 1 |
-Macromolecule #2: Gamma-aminobutyric acid type B receptor subunit 2
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.231258 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASPRSSGQP GPPPPPPPPP ARLLLLLLLP LLLPLAPGAW GWARGAPRPP PSSPPLSIMG LMPLTKEVAK GSIGRGVLPA VELAIEQIR NESLLRPYFL DLRLYDTECD NAKGLKAFYD AIKYGPNHLM VFGGVCPSVT SIIAESLQGW NLVQLSFAAT T PVLADKKK ...String: MASPRSSGQP GPPPPPPPPP ARLLLLLLLP LLLPLAPGAW GWARGAPRPP PSSPPLSIMG LMPLTKEVAK GSIGRGVLPA VELAIEQIR NESLLRPYFL DLRLYDTECD NAKGLKAFYD AIKYGPNHLM VFGGVCPSVT SIIAESLQGW NLVQLSFAAT T PVLADKKK YPYFFRTVPS DNAVNPAILK LLKHYQWKRV GTLTQDVQRF SEVRNDLTGV LYGEDIEISD TESFSNDPCT SV KKLKGND VRIILGQFDQ NMAAKVFCCA YEENMYGSKY QWIIPGWYEP SWWEQVHTEA NSSRCLRKNL LAAMEGYIGV DFE PLSSKQ IKTISGKTPQ QYEREYNNKR SGVGPSKFHG YAYDGIWVIA KTLQRAMETL HASSRHQRIQ DFNYTDHTLG RIIL NAMNE TNFFGVTGQV VFRNGERMGT IKFTQFQDSR EVKVGEYNAV ADTLEIINDT IRFQGSEPPK DKTIILEQLR KISLP LYSI LSALTILGMI MASAFLFFNI KNRNQKLIKM SSPYMNNLII LGGMLSYASI FLFGLDGSFV SEKTFETLCT VRTWIL TVG YTTAFGAMFA KTWRVHAIFK NVKMKKKIIK DQKLLVIVGG MLLIDLCILI CWQAVDPLRR TVEKYSMEPD PAGRDIS IR PLLEHCENTH MTIWLGIVYA YKGLLMLFGC FLAWETRNVS IPALNDSKYI GMSVYNVGIM CIIGAAVSFL TRDQPNVQ F CIVALVIIFC STITLCLVFV PKLITLRTNP DAATQNRRFQ FTQNQKKEDS KTSTSVTSVN QASTSRSGRG GSENLYFQG GSGSGGDYKD DDDKDYKDDD DK UniProtKB: Gamma-aminobutyric acid type B receptor subunit 2 |
-Macromolecule #3: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]...
| Macromolecule | Name: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid type: ligand / ID: 3 / Number of copies: 1 / Formula: 2BV |
|---|---|
| Molecular weight | Theoretical: 408.3 Da |
| Chemical component information | 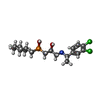 ChemComp-2BV: |
-Macromolecule #4: UNKNOWN LIGAND
| Macromolecule | Name: UNKNOWN LIGAND / type: ligand / ID: 4 / Number of copies: 2 / Formula: UNL |
|---|---|
| Molecular weight | Theoretical: 814.167 Da |
| Chemical component information | 
ChemComp-UNL: |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 16 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Average electron dose: 50.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















