[English] 日本語
 Yorodumi
Yorodumi- EMDB-30301: Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30301 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | |||||||||
 Map data Map data | The focused refinement composite map of the CGP54626-bound GABAB heterodimer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GABAB / Cryo-EM / GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion ...negative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / negative regulation of synaptic transmission / positive regulation of glutamate secretion / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / axolemma / dendritic shaft / response to nicotine / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / mitochondrial membrane / Schaffer collateral - CA1 synapse / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / synaptic vesicle / osteoblast differentiation / transmembrane signaling receptor activity / presynaptic membrane / G alpha (i) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / postsynaptic membrane / neuron projection / G protein-coupled receptor signaling pathway / protein heterodimerization activity / negative regulation of cell population proliferation / neuronal cell body / endoplasmic reticulum membrane / glutamatergic synapse / extracellular space / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Mao C / Shen C | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2020 Journal: Cell Res / Year: 2020Title: Cryo-EM structures of inactive and active GABA receptor. Authors: Chunyou Mao / Cangsong Shen / Chuntao Li / Dan-Dan Shen / Chanjuan Xu / Shenglan Zhang / Rui Zhou / Qingya Shen / Li-Nan Chen / Zhinong Jiang / Jianfeng Liu / Yan Zhang /  Abstract: Metabotropic GABA G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is ...Metabotropic GABA G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is composed of the extracellular Venus flytrap (VFT) domain and transmembrane (TM) domain. Here we present cryo-EM structures of full-length human heterodimeric GABA receptor in the antagonist-bound inactive state and in the active state complexed with an agonist and a positive allosteric modulator in the presence of G protein at a resolution range of 2.8-3.0 Å. Our structures reveal that agonist binding stabilizes the closure of GB1 VFT, which in turn triggers a rearrangement of TM interfaces between the two subunits from TM3-TM5/TM3-TM5 in the inactive state to TM6/TM6 in the active state and finally induces the opening of intracellular loop 3 and synergistic shifting of TM3, 4 and 5 helices in GB2 TM domain to accommodate the α5-helix of G. We also observed that the positive allosteric modulator anchors at the dimeric interface of TM domains. These results provide a structural framework for understanding class C GPCR activation and a rational template for allosteric modulator design targeting the dimeric interface of GABA receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30301.map.gz emd_30301.map.gz | 75.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30301-v30.xml emd-30301-v30.xml emd-30301.xml emd-30301.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30301.png emd_30301.png | 53.4 KB | ||
| Filedesc metadata |  emd-30301.cif.gz emd-30301.cif.gz | 7.1 KB | ||
| Others |  emd_30301_additional.map.gz emd_30301_additional.map.gz | 76.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30301 http://ftp.pdbj.org/pub/emdb/structures/EMD-30301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30301 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30301 | HTTPS FTP |
-Related structure data
| Related structure data |  7c7sMC  7c7qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30301.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30301.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The focused refinement composite map of the CGP54626-bound GABAB heterodimer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: The overall refinement map of the CGP54626-bound GABAB heterodimer.
| File | emd_30301_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The overall refinement map of the CGP54626-bound GABAB heterodimer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The CGP54626-bound GABAB heterodimer
| Entire | Name: The CGP54626-bound GABAB heterodimer |
|---|---|
| Components |
|
-Supramolecule #1: The CGP54626-bound GABAB heterodimer
| Supramolecule | Name: The CGP54626-bound GABAB heterodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gamma-aminobutyric acid type B receptor subunit 1
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 105.976297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFAPGAG GAQTPNATSE GCQIIHPPWE GGIRYRGLTR DQVKAINFLP VDYEIEYVCR GEREVVGPKV RKCLANGSW TDMDTPSRCV RICSKSYLTL ENGKVFLTGG DLPALDGARV DFRCDPDFHL VGSSRSICSQ GQWSTPKPHC Q VNRTPHSE ...String: MKTIIALSYI FCLVFAPGAG GAQTPNATSE GCQIIHPPWE GGIRYRGLTR DQVKAINFLP VDYEIEYVCR GEREVVGPKV RKCLANGSW TDMDTPSRCV RICSKSYLTL ENGKVFLTGG DLPALDGARV DFRCDPDFHL VGSSRSICSQ GQWSTPKPHC Q VNRTPHSE RRAVYIGALF PMSGGWPGGQ ACQPAVEMAL EDVNSRRDIL PDYELKLIHH DSKCDPGQAT KYLYELLYND PI KIILMPG CSSVSTLVAE AARMWNLIVL SYGSSSPALS NRQRFPTFFR THPSATLHNP TRVKLFEKWG WKKIATIQQT TEV FTSTLD DLEERVKEAG IEITFRQSFF SDPAVPVKNL KRQDARIIVG LFYETEARKV FCEVYKERLF GKKYVWFLIG WYAD NWFKI YDPSINCTVD EMTEAVEGHI TTEIVMLNPA NTRSISNMTS QEFVEKLTKR LKRHPEETGG FQEAPLAYDA IWALA LALN KTSGGGGRSG VRLEDFNYNN QTITDQIYRA MNSSSFEGVS GHVVFDASGS RMAWTLIEQL QGGSYKKIGY YDSTKD DLS WSKTDKWIGG SPPADQTLVI KTFRFLSQKL FISVSVLSSL GIVLAVVCLS FNIYNSHVRY IQNSQPNLNN LTAVGCS LA LAAVFPLGLD GYHIGRNQFP FVCQARLWLL GLGFSLGYGS MFTKIWWVHT VFTKKEEKKE WRKTLEPWKL YATVGLLV G MDVLTLAIWQ IVDPLHRTIE TFAKEEPKED IDVSILPQLE HCSSRKMNTW LGIFYGYKGL LLLLGIFLAY ETKSVSTEK INDHRAVGMA IYNVAVLCLI TAPVTMILSS QQDAAFAFAS LAIVFSSYIT LVVLFVPKMR RLITRGEWQS EAQDTMKTGS STNNNEEEK SRLLEKENRE LEKIIAEKEE RVSELRHQLQ SRLEVLFQGP HHHHHHHH UniProtKB: Gamma-aminobutyric acid type B receptor subunit 1 |
-Macromolecule #2: Gamma-aminobutyric acid type B receptor subunit 2
| Macromolecule | Name: Gamma-aminobutyric acid type B receptor subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 91.359445 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKGSGGSG WARGAPRPPP SSPPLSIMGL MPLTKEVAKG SIGRGVLPAV ELAIEQIRNE SLLRPYFLD LRLYDTECDN AKGLKAFYDA IKYGPNHLMV FGGVCPSVTS IIAESLQGWN LVQLSFAATT PVLADKKKYP Y FFRTVPSD ...String: MKTIIALSYI FCLVFADYKD DDDKGSGGSG WARGAPRPPP SSPPLSIMGL MPLTKEVAKG SIGRGVLPAV ELAIEQIRNE SLLRPYFLD LRLYDTECDN AKGLKAFYDA IKYGPNHLMV FGGVCPSVTS IIAESLQGWN LVQLSFAATT PVLADKKKYP Y FFRTVPSD NAVNPAILKL LKHYQWKRVG TLTQDVQRFS EVRNDLTGVL YGEDIEISDT ESFSNDPCTS VKKLKGNDVR II LGQFDQN MAAKVFCCAY EENMYGSKYQ WIIPGWYEPS WWEQVHTEAN SSRCLRKNLL AAMEGYIGVD FEPLSSKQIK TIS GKTPQQ YEREYNNKRS GVGPSKFHGY AYDGIWVIAK TLQRAMETLH ASSRHQRIQD FNYTDHTLGR IILNAMNETN FFGV TGQVV FRNGERMGTI KFTQFQDSRE VKVGEYNAVA DTLEIINDTI RFQGSEPPKD KTIILEQLRK ISLPLYSILS ALTIL GMIM ASAFLFFNIK NRNQKLIKMS SPYMNNLIIL GGMLSYASIF LFGLDGSFVS EKTFETLCTV RTWILTVGYT TAFGAM FAK TWRVHAIFKN VKMKKKIIKD QKLLVIVGGM LLIDLCILIC WQAVDPLRRT VEKYSMEPDP AGRDISIRPL LEHCENT HM TIWLGIVYAY KGLLMLFGCF LAWETRNVSI PALNDSKYIG MSVYNVGIMC IIGAAVSFLT RDQPNVQFCI VALVIIFC S TITLCLVFVP KLITLRTNPD AATQNRRFQF TQNQKKEDSK TSTSVTSVNQ ASTSRLEGLQ SENHRLRMKI TELDKDLEE VTMQLQDT UniProtKB: Gamma-aminobutyric acid type B receptor subunit 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]...
| Macromolecule | Name: (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid type: ligand / ID: 4 / Number of copies: 1 / Formula: 2BV |
|---|---|
| Molecular weight | Theoretical: 408.3 Da |
| Chemical component information | 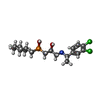 ChemComp-2BV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number real images: 4740 / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49310 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






























