[English] 日本語
 Yorodumi
Yorodumi- PDB-7c7q: Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) recept... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7c7q | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / GABAB / Cryo-EM / GPCR / active / PAM | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion ...negative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / : / negative regulation of epinephrine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / negative regulation of synaptic transmission / positive regulation of glutamate secretion / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / axolemma / dendritic shaft / response to nicotine / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / mitochondrial membrane / Schaffer collateral - CA1 synapse / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / synaptic vesicle / osteoblast differentiation / transmembrane signaling receptor activity / presynaptic membrane / G alpha (i) signalling events / dendritic spine / response to ethanol / chemical synaptic transmission / postsynaptic membrane / neuron projection / G protein-coupled receptor signaling pathway / protein heterodimerization activity / negative regulation of cell population proliferation / neuronal cell body / endoplasmic reticulum membrane / glutamatergic synapse / extracellular space / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3 Å | |||||||||
 Authors Authors | Mao, C. / Shen, C. / Li, C. / Shen, D. / Xu, C. / Zhang, S. / Zhou, R. / Shen, Q. / Chen, L. / Jiang, Z. ...Mao, C. / Shen, C. / Li, C. / Shen, D. / Xu, C. / Zhang, S. / Zhou, R. / Shen, Q. / Chen, L. / Jiang, Z. / Liu, J. / Zhang, Y. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2020 Journal: Cell Res / Year: 2020Title: Cryo-EM structures of inactive and active GABA receptor. Authors: Chunyou Mao / Cangsong Shen / Chuntao Li / Dan-Dan Shen / Chanjuan Xu / Shenglan Zhang / Rui Zhou / Qingya Shen / Li-Nan Chen / Zhinong Jiang / Jianfeng Liu / Yan Zhang /  Abstract: Metabotropic GABA G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is ...Metabotropic GABA G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is composed of the extracellular Venus flytrap (VFT) domain and transmembrane (TM) domain. Here we present cryo-EM structures of full-length human heterodimeric GABA receptor in the antagonist-bound inactive state and in the active state complexed with an agonist and a positive allosteric modulator in the presence of G protein at a resolution range of 2.8-3.0 Å. Our structures reveal that agonist binding stabilizes the closure of GB1 VFT, which in turn triggers a rearrangement of TM interfaces between the two subunits from TM3-TM5/TM3-TM5 in the inactive state to TM6/TM6 in the active state and finally induces the opening of intracellular loop 3 and synergistic shifting of TM3, 4 and 5 helices in GB2 TM domain to accommodate the α5-helix of G. We also observed that the positive allosteric modulator anchors at the dimeric interface of TM domains. These results provide a structural framework for understanding class C GPCR activation and a rational template for allosteric modulator design targeting the dimeric interface of GABA receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7c7q.cif.gz 7c7q.cif.gz | 261.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7c7q.ent.gz pdb7c7q.ent.gz | 201.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7c7q.json.gz 7c7q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c7/7c7q https://data.pdbj.org/pub/pdb/validation_reports/c7/7c7q ftp://data.pdbj.org/pub/pdb/validation_reports/c7/7c7q ftp://data.pdbj.org/pub/pdb/validation_reports/c7/7c7q | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  30300MC  7c7sC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 99082.711 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR1, GPRC3A / Cell line (production host): HEK 293F / Production host: Homo sapiens (human) / Gene: GABBR1, GPRC3A / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human) / References: UniProt: Q9UBS5 Homo sapiens (human) / References: UniProt: Q9UBS5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Protein | Mass: 86812.398 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Cell line (production host): HEK 293F / Production host: Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Cell line (production host): HEK 293F / Production host:  Homo sapiens (human) / References: UniProt: O75899 Homo sapiens (human) / References: UniProt: O75899 | ||||||||
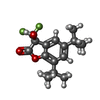
| #3: Sugar | ChemComp-NAG / #4: Chemical | ChemComp-FN0 / ( | #5: Chemical | ChemComp-2C0 / | Has ligand of interest | Y | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: The baclofen/BHFF-bound GABAB heterodimer / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: HEK 293F Homo sapiens (human) / Cell: HEK 293F | ||||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||||
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 29000 X / Calibrated magnification: 49310 X / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 64 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 4624 |
| Image scans | Movie frames/image: 40 / Used frames/image: 1-40 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.16_3549: / Classification: refinement | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 3075533 | ||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 237606 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 7C7S Accession code: 7C7S / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj