[English] 日本語
 Yorodumi
Yorodumi- EMDB-10793: Cryo-EM structure of the Full-length disease type human Huntingtin -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10793 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Full-length disease type human Huntingtin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | multivalent scaffold platform / PROTEIN BINDING / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / vesicle transport along microtubule ...: / positive regulation of CAMKK-AMPK signaling cascade / microtubule-based transport / vocal learning / regulation of CAMKK-AMPK signaling cascade / positive regulation of mitophagy / profilin binding / positive regulation of cilium assembly / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / vesicle transport along microtubule / positive regulation of aggrephagy / positive regulation of lipophagy / Golgi organization / dynein intermediate chain binding / dynactin binding / establishment of mitotic spindle orientation / Regulation of MECP2 expression and activity / postsynaptic cytosol / beta-tubulin binding / presynaptic cytosol / heat shock protein binding / phosphoprotein phosphatase activity / inclusion body / centriole / autophagosome / cytoplasmic vesicle membrane / negative regulation of extrinsic apoptotic signaling pathway / protein destabilization / kinase binding / p53 binding / late endosome / transmembrane transporter binding / early endosome / positive regulation of apoptotic process / axon / apoptotic process / dendrite / perinuclear region of cytoplasm / endoplasmic reticulum / Golgi apparatus / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
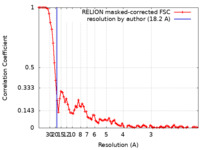
| Method | single particle reconstruction / cryo EM / Resolution: 18.2 Å | |||||||||
 Authors Authors | Jung T / Tamo G / Dal Perraro M / Hebert H / Song J | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain. Authors: Taeyang Jung / Baehyun Shin / Giorgio Tamo / Hyeongju Kim / Ravi Vijayvargia / Alexander Leitner / Maria J Marcaida / Juan Astorga-Wells / Roy Jung / Ruedi Aebersold / Matteo Dal Peraro / ...Authors: Taeyang Jung / Baehyun Shin / Giorgio Tamo / Hyeongju Kim / Ravi Vijayvargia / Alexander Leitner / Maria J Marcaida / Juan Astorga-Wells / Roy Jung / Ruedi Aebersold / Matteo Dal Peraro / Hans Hebert / Ihn Sik Seong / Ji-Joon Song /     Abstract: The polyQ expansion in huntingtin protein (HTT) is the prime cause of Huntington's disease (HD). The recent cryoelectron microscopy (cryo-EM) structure of HTT-HAP40 complex provided the structural ...The polyQ expansion in huntingtin protein (HTT) is the prime cause of Huntington's disease (HD). The recent cryoelectron microscopy (cryo-EM) structure of HTT-HAP40 complex provided the structural information on its HEAT-repeat domains. Here, we present analyses of the impact of polyQ length on the structure and function of HTT via an integrative structural and biochemical approach. The cryo-EM analysis of normal (Q23) and disease (Q78) type HTTs shows that the structures of apo HTTs significantly differ from the structure of HTT in a HAP40 complex and that the polyQ expansion induces global structural changes in the relative movements among the HTT domains. In addition, we show that the polyQ expansion alters the phosphorylation pattern across HTT and that Ser2116 phosphorylation in turn affects the global structure and function of HTT. These results provide a molecular basis for the effect of the polyQ segment on HTT structure and activity, which may be important for HTT pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10793.map.gz emd_10793.map.gz | 19.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10793-v30.xml emd-10793-v30.xml emd-10793.xml emd-10793.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10793_fsc.xml emd_10793_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_10793.png emd_10793.png | 41 KB | ||
| Filedesc metadata |  emd-10793.cif.gz emd-10793.cif.gz | 8.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10793 http://ftp.pdbj.org/pub/emdb/structures/EMD-10793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10793 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10793 | HTTPS FTP |
-Related structure data
| Related structure data |  6yejMC  4937C  4944C  6rmhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10793.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10793.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Full-length human disease-type huntingtin
| Entire | Name: Full-length human disease-type huntingtin |
|---|---|
| Components |
|
-Supramolecule #1: Full-length human disease-type huntingtin
| Supramolecule | Name: Full-length human disease-type huntingtin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: A disease type huntingtin with 78 polyglutamine repeat tract |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Huntingtin
| Macromolecule | Name: Huntingtin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 355.278719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQ QQQQQQPPPP PPPPPPPQLP QPPPQAQPLL PQPQPPPPPP PPPPGPAVAE EPLHRPKKEL SATKKDRVNH C LTICENIV ...String: MATLEKLMKA FESLKSFQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQQ QQQQQQQQQ QQQQQQPPPP PPPPPPPQLP QPPPQAQPLL PQPQPPPPPP PPPPGPAVAE EPLHRPKKEL SATKKDRVNH C LTICENIV AQSVRNSPEF QKLLGIAMEL FLLCSDDAES DVRMVADECL NKVIKALMDS NLPRLQLELY KEIKKNGAPR SL RAALWRF AELAHLVRPQ KCRPYLVNLL PCLTRTSKRP EESVQETLAA AVPKIMASFG NFANDNEIKV LLKAFIANLK SSS PTIRRT AAGSAVSICQ HSRRTQYFYS WLLNVLLGLL VPVEDEHSTL LILGVLLTLR YLVPLLQQQV KDTSLKGSFG VTRK EMEVS PSAEQLVQVY ELTLHHTQHQ DHNVVTGALE LLQQLFRTPP PELLQTLTAV GGIGQLTAAK EESGGRSRSG SIVEL IAGG GSSCSPVLSR KQKGKVLLGE EEALEDDSES RSDVSSSALT ASVKDEISGE LAASSGVSTP GSAGHDIITE QPRSQH TLQ ADSVDLASCD LTSSATDGDE EDILSHSSSQ VSAVPSDPAM DLNDGTQASS PISDSSQTTT EGPDSAVTPS DSSEIVL DG TDNQYLGLQI GQPQDEDEEA TGILPDEASE AFRNSSMALQ QAHLLKNMSH CRQPSDSSVD KFVLRDEATE PGDQENKP C RIKGDIGQST DDDSAPLVHC VRLLSASFLL TGGKNVLVPD RDVRVSVKAL ALSCVGAAVA LHPESFFSKL YKVPLDTTE YPEEQYVSDI LNYIDHGDPQ VRGATAILCG TLICSILSRS RFHVGDWMGT IRTLTGNTFS LADCIPLLRK TLKDESSVTC KLACTAVRN CVMSLCSSSY SELGLQLIID VLTLRNSSYW LVRTELLETL AEIDFRLVSF LEAKAENLHR GAHHYTGLLK L QERVLNNV VIHLLGDEDP RVRHVAAASL IRLVPKLFYK CDQGQADPVV AVARDQSSVY LKLLMHETQP PSHFSVSTIT RI YRGYNLL PSITDVTMEN NLSRVIAAVS HELITSTTRA LTFGCCEALC LLSTAFPVCI WSLGWHCGVP PLSASDESRK SCT VGMATM ILTLLSSAWF PLDLSAHQDA LILAGNLLAA SAPKSLRSSW ASEEEANPAA TKQEEVWPAL GDRALVPMVE QLFS HLLKV INICAHVLDD VAPGPAIKAA LPSLTNPPSL SPIRRKGKEK EPGEQASVPL SPKKGSEASA ASRQSDTSGP VTTSK SSSL GSFYHLPSYL RLHDVLKATH ANYKVTLDLQ NSTEKFGGFL RSALDVLSQI LELATLQDIG KCVEEILGYL KSCFSR EPM MATVCVQQLL KTLFGTNLAS QFDGLSSNPS KSQGRAQRLG SSSVRPGLYH YCFMAPYTHF TQALADASLR NMVQAEQ EN DTSGWFDVLQ KVSTQLKTNL TSVTKNRADK NAIHNHIRLF EPLVIKALKQ YTTTTCVQLQ KQVLDLLAQL VQLRVNYC L LDSDQVFIGF VLKQFEYIEV GQFRESEAII PNIFFFLVLL SYERYHSKQI IGIPKIIQLC DGIMASGRKA VTHAIPALQ PIVHDLFVLR GTNKADAGKE LETQKEVVVS MLLRLIQYHQ VLEMFILVLQ QCHKENEDKW KRLSRQIADI ILPMLAKQQM HIDSHEALG VLNTLFEILA PSSLRPVDML LRSMFVTPNT MASVSTVQLW ISGILAILRV LISQSTEDIV LSRIQELSFS P YLISCTVI NRLRDGDSTS TLEEHSEGKQ IKNLPEETFS RFLLQLVGIL LEDIVTKQLK VEMSEQQHTF YCQELGTLLM CL IHIFKSG MFRRITAAAT RLFRSDGCGG SFYTLDSLNL RARSMITTHP ALVLLWCQIL LLVNHTDYRW WAEVQQTPKR HSL SSTKLL SPQMSGEEED SDLAAKLGMC NREIVRRGAL ILFCDYVCQN LHDSEHLTWL IVNHIQDLIS LSHEPPVQDF ISAV HRNSA ASGLFIQAIQ SRCENLSTPT MLKKTLQCLE GIHLSQSGAV LTLYVDRLLC TPFRVLARMV DILACRRVEM LLAAN LQSS MAQLPMEELN RIQEYLQSSG LAQRHQRLYS LLDRFRLSTM QDSLSPSPPV SSHPLDGDGH VSLETVSPDK DWYVHL VKS QCWTRSDSAL LEGAELVNRI PAEDMNAFMM NSEFNLSLLA PCLSLGMSEI SGGQKSALFE AAREVTLARV SGTVQQL PA VHHVFQPELP AEPAAYWSKL NDLFGDAALY QSLPTLARAL AQYLVVVSKL PSHLHLPPEK EKDIVKFVVA TLEALSWH L IHEQIPLSLD LQAGLDCCCL ALQLPGLWSV VSSTEFVTHA CSLIHCVHFI LEAVAVQPGE QLLSPERRTN TPKAISEEE EEVDPNTQNP KYITAACEMV AEMVESLQSV LALGHKRNSG VPAFLTPLLR NIIISLARLP LVNSYTRVPP LVWKLGWSPK PGGDFGTAF PEIPVEFLQE KEVFKEFIYR INTLGWTSRT QFEETWATLL GVLVTQPLVM EQEESPPEED TERTQINVLA V QAITSLVL SAMTVPVAGN PAVSCLEQQP RNKPLKALDT RFGRKLSIIR GIVEQEIQAM VSKRENIATH HLYQAWDPVP SL SPATTGA LISHEKLLLQ INPERELGSM SYKLGQVSIH SVWLGNSITP LREEEWDEEE EEEADAPAPS SPPTSPVNSR KHR AGVDIH SCSQFLLELY SRWILPSSSA RRTPAILISE VVRSLLVVSD LFTERNQFEL MYVTLTELRR VHPSEDEILA QYLV PATCK AAAVLGMDKA VAEPVSRLLE STLRSSHLPS RVGALHGVLY VLECDLLDDT AKQLIPVISD YLLSNLKGIA HCVNI HSQQ HVLVMCATAF YLIENYPLDV GPEFSASIIQ MCGVMLSGSE ESTPSIIYHC ALRGLERLLL SEQLSRLDAE SLVKLS VDR VNVHSPHRAM AALGLMLTCM YTGKEKVSPG RTSDPNPAAP DSESVIVAME RVSVLFDRIR KGFPCEARVV ARILPQF LD DFFPPQDIMN KVIGEFLSNQ QPYPQFMATV VYKVFQTLHS TGQSSMVRDW VMLSLSNFTQ RAPVAMATWS LSCFFVSA S TSPWVAAILP HVISRMGKLE QVDVNLFCLV ATDFYRHQIE EELDRRAFQS VLEVVAAPGS PYHRLLTCLR NVHKVTTC UniProtKB: Huntingtin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.08 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK II / Details: 30 seconds incubation 8 seconds blotting. | |||||||||
| Details | HTT was mixed with final 0.05% of Octyl glucoside. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-24 / Number real images: 1500 / Average exposure time: 6.0 sec. / Average electron dose: 33.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47170 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.4 µm / Nominal defocus min: 0.4 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)