-Search query
-Search result
Showing all 50 items for (author: calcraft & t)

EMDB-49737: 
Subtomogram Average of in situ Lipoprotein Lipase Filament
Method: subtomogram averaging / : Gunn KH, Wheless A, Neher SB

EMDB-49738: 
Lipoprotein Lipase Helical Filament with 11 nm diameter
Method: helical / : Gunn KH, Wheless A, Neher SB

PDB-9nrn: 
Lipoprotein Lipase Helical Filament with 11 nm diameter
Method: helical / : Gunn KH, Wheless A, Neher SB

PDB-9hmf: 
Periplasmic scaffold of the Campylobacter jejuni flagellar motor (alpha carbon trace)
Method: single particle / : Drobnic T, Beeby M

EMDB-17415: 
Campylobacter jejuni flagellar motor, pflC deletion
Method: subtomogram averaging / : Drobnic T, Alzheimer M, Svensson S, Sharma CS, Beeby M

EMDB-17416: 
Campylobacter jejuni flagellar motor, pflD deletion
Method: subtomogram averaging / : Drobnic T, Henderson LD, Alzheimer M, Svensson S, Sharma CM, Beeby M

EMDB-17417: 
Campylobacter jejuni flagellar motor, truncated PflA (d16-168)
Method: subtomogram averaging / : Drobnic T, Nans A, Rosenthal PB, Beeby M

EMDB-17419: 
Campylobacter jejuni flagellar motor, FlgQ-mCherry fusion
Method: subtomogram averaging / : Drobnic T, Hendrixson DR, Beeby M

EMDB-19642: 
Campylobacter jejuni bacterial flagellar C-ring
Method: subtomogram averaging / : Beeby M

EMDB-16723: 
Wild-type Campylobacter jejuni flagellar motor, in situ
Method: single particle / : Drobnic T, Cohen EJ, Calcraft T, Singh NK, Nans A, Rosenthal PB, Beeby M

EMDB-16724: 
Periplasmic scaffold of the Campylobacter jejuni flagellar motor
Method: single particle / : Drobnic T, Cohen EJ, Singh NK, Umrekar TR, Nans A, Rosenthal PB, Beeby M

EMDB-17309: 
In situ cryoEM structure of Prototype Foamy Virus Env trimer
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17311: 
In situ cryoEM structure of Prototype Foamy Virus Env dimer of trimers
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17312: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, icosahedral map
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17313: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, pentamer localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17314: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, hexamer 1 localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17315: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, hexamer 2 localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

EMDB-17316: 
In situ subtomogram average of Prototype Foamy Virus Env trimer
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-17317: 
In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-17318: 
In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-17319: 
In situ subtomogram average of the Prototype Foamy Virus capsid, wild-type Gag
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-17320: 
In situ subtomogram average of the Prototype Foamy Virus capsid, p68 Gag
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-17321: 
Cryotomogram of Prototype Foamy Virus particles, wild-type Gag
Method: electron tomography / : Calcraft T, Nans A, Rosenthal PB
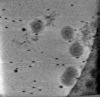
EMDB-17322: 
Cryotomogram of Prototype Foamy Virus particles, p68 Gag
Method: electron tomography / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozh: 
In situ cryoEM structure of Prototype Foamy Virus Env trimer
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozj: 
In situ cryoEM structure of Prototype Foamy Virus Env dimer of trimers
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozk: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, icosahedral map
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozl: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, pentamer localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozm: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, hexamer 1 localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozn: 
In situ cryoEM structure of the Prototype Foamy Virus capsid, hexamer 2 localised reconstruction
Method: single particle / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozp: 
In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

PDB-8ozq: 
In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers
Method: subtomogram averaging / : Calcraft T, Nans A, Rosenthal PB

EMDB-18916: 
Cryotomogram of mature Vaccinia virus (WR) virion
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18917: 
Subtomogram average of the Vaccinia virus (WR) portal complex in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18918: 
Subtomogram average of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8r5i: 
In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15596: 
In situ subtomogram average of Vaccinia virus (WR) palisade, all virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15597: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from CEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15598: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IMVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15599: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15600: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (immature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15601: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15602: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8arh: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15181: 
Prefusion spike of SARS-CoV-2 (Wuhan), closed conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB
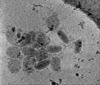
EMDB-15182: 
Electron cryotomography of SARS-CoV-2 virions
Method: electron tomography / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15183: 
Whole SARS-CoV-2 (Wuhan) virion
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15185: 
Prefusion spike of SARS-CoV-2 (Wuhan), 1-RBD-up conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-11928: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T

PDB-7awa: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, Grabe G, Holden D, Nans A, Rosenthal P, Rouse S, Beeby M
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
