+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
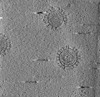
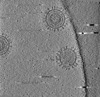
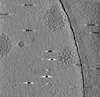
| Title | Cryotomogram of Prototype Foamy Virus particles, wild-type Gag | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | capsid / Gag / full-length / CA domain / VIRAL PROTEIN | |||||||||
| Biological species |  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Calcraft T / Nans A / Rosenthal PB | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry. Authors: Thomas Calcraft / Nicole Stanke-Scheffler / Andrea Nans / Dirk Lindemann / Ian A Taylor / Peter B Rosenthal /   Abstract: Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates ...Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates for gene and oncolytic therapies. Structural studies of PFV contribute to the understanding of the mechanisms of FV replication, cell entry and infection, and retroviral evolution. Here we combine cryoEM and cryoET to determine high-resolution in situ structures of the PFV icosahedral capsid (CA) and envelope glycoprotein (Env), including its type III transmembrane anchor and membrane-proximal external region (MPER), and show how they are organized in an integrated structure of assembled PFV particles. The atomic models reveal an ancient retroviral capsid architecture and an unexpected relationship between Env and other class 1 fusion proteins of the Mononegavirales. Our results represent the de novo structure determination of an assembled retrovirus particle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17321.map.gz emd_17321.map.gz | 965.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17321-v30.xml emd-17321-v30.xml emd-17321.xml emd-17321.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17321.png emd_17321.png | 220.3 KB | ||
| Filedesc metadata |  emd-17321.cif.gz emd-17321.cif.gz | 4.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17321 http://ftp.pdbj.org/pub/emdb/structures/EMD-17321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17321 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17321.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17321.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.52 Å | ||||||||||||||||||||||||||||||||
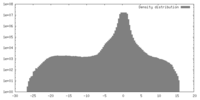
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Eastern chimpanzee simian foamy virus
| Entire | Name:  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern chimpanzee simian foamy virus
| Supramolecule | Name: Eastern chimpanzee simian foamy virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 2170195 / Sci species name: Eastern chimpanzee simian foamy virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: BBI Solutions / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION Software:
Number images used: 41 |
|---|
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)