[English] 日本語
 Yorodumi
Yorodumi- EMDB-17316: In situ subtomogram average of Prototype Foamy Virus Env trimer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ subtomogram average of Prototype Foamy Virus Env trimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane fusion / envelope glycoprotein / foamy virus / MPER / transmembrane / MEMBRANE PROTEIN / VIRAL PROTEIN | |||||||||
| Biological species |  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus | |||||||||
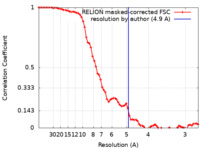
| Method | subtomogram averaging / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Calcraft T / Nans A / Rosenthal PB | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry. Authors: Thomas Calcraft / Nicole Stanke-Scheffler / Andrea Nans / Dirk Lindemann / Ian A Taylor / Peter B Rosenthal /   Abstract: Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates ...Foamy viruses (FVs) are an ancient lineage of retroviruses, with an evolutionary history spanning over 450 million years. Vector systems based on Prototype Foamy Virus (PFV) are promising candidates for gene and oncolytic therapies. Structural studies of PFV contribute to the understanding of the mechanisms of FV replication, cell entry and infection, and retroviral evolution. Here we combine cryoEM and cryoET to determine high-resolution in situ structures of the PFV icosahedral capsid (CA) and envelope glycoprotein (Env), including its type III transmembrane anchor and membrane-proximal external region (MPER), and show how they are organized in an integrated structure of assembled PFV particles. The atomic models reveal an ancient retroviral capsid architecture and an unexpected relationship between Env and other class 1 fusion proteins of the Mononegavirales. Our results represent the de novo structure determination of an assembled retrovirus particle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17316.map.gz emd_17316.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17316-v30.xml emd-17316-v30.xml emd-17316.xml emd-17316.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17316_fsc.xml emd_17316_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17316.png emd_17316.png | 55.1 KB | ||
| Masks |  emd_17316_msk_1.map emd_17316_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17316.cif.gz emd-17316.cif.gz | 5.9 KB | ||
| Others |  emd_17316_half_map_1.map.gz emd_17316_half_map_1.map.gz emd_17316_half_map_2.map.gz emd_17316_half_map_2.map.gz | 15.1 MB 15.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17316 http://ftp.pdbj.org/pub/emdb/structures/EMD-17316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17316 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17316 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17316.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17316.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17316_msk_1.map emd_17316_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17316_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17316_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern chimpanzee simian foamy virus
| Entire | Name:  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern chimpanzee simian foamy virus
| Supramolecule | Name: Eastern chimpanzee simian foamy virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2170195 / Sci species name: Eastern chimpanzee simian foamy virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Envelope glycoprotein
| Macromolecule | Name: Envelope glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Eastern chimpanzee simian foamy virus Eastern chimpanzee simian foamy virus |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAPPMTLQQW IIWNKMNKAH EALQNTTTVT EQQKEQIILD IQNEEVQPTR RDKFRYLLYT CCATSSRVLA WIFLVCILLI IVLVSCFVTI SRIQWNKDIQ VLGPVIDWNV TQRAVYQPLQ TRRIARSLRM QHPVPKYVEV NMTSIPQGVY YEPHPEPIVV KERVLGLSQI ...String: MAPPMTLQQW IIWNKMNKAH EALQNTTTVT EQQKEQIILD IQNEEVQPTR RDKFRYLLYT CCATSSRVLA WIFLVCILLI IVLVSCFVTI SRIQWNKDIQ VLGPVIDWNV TQRAVYQPLQ TRRIARSLRM QHPVPKYVEV NMTSIPQGVY YEPHPEPIVV KERVLGLSQI LMINSENIAN NANLTQEVKK LLTEMVNEEM QSLSDVMIDF EIPLGDPRDQ EQYIHRKCYQ EFANCYLVKY KEPKPWPKEG LIADQCPLPG YHAGLTYNRQ SIWDYYIKVE SIRPANWTTK SKYGQARLGS FYIPSSLRQI NVSHVLFCSD QLYSKWYNIE NTIEQNERFL LNKLNNLTSG TSVLKKRALP KDWSSQGKNA LFREINVLDI CSKPESVILL NTSYYSFSLW EGDCNFTKDM ISQLVPECDG FYNNSKWMHM HPYACRFWRS KNEKEETKCR DGETKRCLYY PLWDSPESTY DFGYLAYQKN FPSPICIEQQ KIRDQDYEVY SLYQECKIAS KAYGIDTVLF SLKNFLNYTG TPVNEMPNAR AFVGLIDPKF PPSYPNVTRE HYTSCNNRKR RSVDNNYAKL RSMGYALTGA VQTLSQISDI NDENLQQGIY LLRDHVITLM EATLHDISVM EGMFAVQHLH THLNHLKTML LERRIDWTYM SSTWLQQQLQ KSDDEMKVIK RIARSLVYYV KQTHSSPTAT AWEIGLYYEL VIPKHIYLNN WNVVNIGHLV KSAGQLTHVT IAHPYEIINK ECVETIYLHL EDCTRQDYVI CDVVKIVQPC GNSSDTSDCP VWAEAVKEPF VQVNPLKNGS YLVLASSTDC QIPPYVPSIV TVNETTSCFG LDFKRPLVAE ERLSFEPRLP NLQLRLPHLV GIIAKIKGIK IEVTSSGESI KEQIERAKAE LLRLDIHEGD TPAWIQQLAA ATKDVWPAAA SALQGIGNFL SGTAQGIFGT AFSLLGYLKP ILIGVGVILL VILIFKIVSW IPTKKKNQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)