[English] 日本語
 Yorodumi
Yorodumi- EMDB-15601: Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature vi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
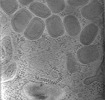
| Title | Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature virions) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Palisade / Lattice / A4 / VIRUS | |||||||||
| Biological species |  Vaccinia virus WR Vaccinia virus WR | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Calcraft T / Hernandez-Gonzalez M / Nans A / Rosenthal PB / Way M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: A succession of two viral lattices drives vaccinia virus assembly. Authors: Miguel Hernandez-Gonzalez / Thomas Calcraft / Andrea Nans / Peter B Rosenthal / Michael Way /  Abstract: During its cytoplasmic replication, vaccinia virus assembles non-infectious spherical immature virions (IV) coated by a viral D13 lattice. Subsequently, IV mature into infectious brick-shaped ...During its cytoplasmic replication, vaccinia virus assembles non-infectious spherical immature virions (IV) coated by a viral D13 lattice. Subsequently, IV mature into infectious brick-shaped intracellular mature virions (IMV) that lack D13. Here, we performed cryo-electron tomography (cryo-ET) of frozen-hydrated vaccinia-infected cells to structurally characterise the maturation process in situ. During IMV formation, a new viral core forms inside IV with a wall consisting of trimeric pillars arranged in a new pseudohexagonal lattice. This lattice appears as a palisade in cross-section. As maturation occurs, which involves a 50% reduction in particle volume, the viral membrane becomes corrugated as it adapts to the newly formed viral core in a process that does not appear to require membrane removal. Our study suggests that the length of this core is determined by the D13 lattice and that the consecutive D13 and palisade lattices control virion shape and dimensions during vaccinia assembly and maturation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15601.map.gz emd_15601.map.gz | 41.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15601-v30.xml emd-15601-v30.xml emd-15601.xml emd-15601.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15601.png emd_15601.png | 166.8 KB | ||
| Filedesc metadata |  emd-15601.cif.gz emd-15601.cif.gz | 4.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15601 http://ftp.pdbj.org/pub/emdb/structures/EMD-15601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15601 | HTTPS FTP |
-Validation report
| Summary document |  emd_15601_validation.pdf.gz emd_15601_validation.pdf.gz | 560.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_15601_full_validation.pdf.gz emd_15601_full_validation.pdf.gz | 559.6 KB | Display | |
| Data in XML |  emd_15601_validation.xml.gz emd_15601_validation.xml.gz | 5.1 KB | Display | |
| Data in CIF |  emd_15601_validation.cif.gz emd_15601_validation.cif.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15601 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15601 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15601 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-15601 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15601.map.gz / Format: CCP4 / Size: 51.4 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_15601.map.gz / Format: CCP4 / Size: 51.4 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 25.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Vaccinia virus WR
| Entire | Name:  Vaccinia virus WR Vaccinia virus WR |
|---|---|
| Components |
|
-Supramolecule #1: Vaccinia virus WR
| Supramolecule | Name: Vaccinia virus WR / type: virus / ID: 1 / Parent: 0 / Details: A36-YdF deltaF11 / NCBI-ID: 10254 / Sci species name: Vaccinia virus WR / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: BBI / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 8.0 µm / Nominal magnification: 11600 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 39 IMOD / Number images used: 39 |
|---|
 Movie
Movie Controller
Controller