+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11928 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SctV (SsaV) cytoplasmic domain | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Type III secretion / T3SS / Export Apparatus / PROTEIN TRANSPORT | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Salmonella enterica (bacteria) / Salmonella enterica (bacteria) /  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) | |||||||||||||||
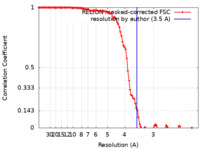
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Matthews-Palmer TRS / Gonzalez-Rodriguez N / Calcraft T / Lagercrantz S / Zachs T | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2021 Journal: J Struct Biol / Year: 2021Title: Structure of the cytoplasmic domain of SctV (SsaV) from the Salmonella SPI-2 injectisome and implications for a pH sensing mechanism. Authors: Teige R S Matthews-Palmer / Nayim Gonzalez-Rodriguez / Thomas Calcraft / Signe Lagercrantz / Tobias Zachs / Xiu-Jun Yu / Grzegorz J Grabe / David W Holden / Andrea Nans / Peter B Rosenthal / ...Authors: Teige R S Matthews-Palmer / Nayim Gonzalez-Rodriguez / Thomas Calcraft / Signe Lagercrantz / Tobias Zachs / Xiu-Jun Yu / Grzegorz J Grabe / David W Holden / Andrea Nans / Peter B Rosenthal / Sarah L Rouse / Morgan Beeby /  Abstract: Bacterial type III secretion systems assemble the axial structures of both injectisomes and flagella. Injectisome type III secretion systems subsequently secrete effector proteins through their ...Bacterial type III secretion systems assemble the axial structures of both injectisomes and flagella. Injectisome type III secretion systems subsequently secrete effector proteins through their hollow needle into a host, requiring co-ordination. In the Salmonella enterica serovar Typhimurium SPI-2 injectisome, this switch is triggered by sensing the neutral pH of the host cytoplasm. Central to specificity switching is a nonameric SctV protein with an N-terminal transmembrane domain and a toroidal C-terminal cytoplasmic domain. A 'gatekeeper' complex interacts with the SctV cytoplasmic domain in a pH dependent manner, facilitating translocon secretion while repressing effector secretion through a poorly understood mechanism. To better understand the role of SctV in SPI-2 translocon-effector specificity switching, we purified full-length SctV and determined its toroidal cytoplasmic region's structure using cryo-EM. Structural comparisons and molecular dynamics simulations revealed that the cytoplasmic torus is stabilized by its core subdomain 3, about which subdomains 2 and 4 hinge, varying the flexible outside cleft implicated in gatekeeper and substrate binding. In light of patterns of surface conservation, deprotonation, and structural motion, the location of previously identified critical residues suggest that gatekeeper binds a cleft buried between neighboring subdomain 4s. Simulations suggest that a local pH change from 5 to 7.2 stabilizes the subdomain 3 hinge and narrows the central aperture of the nonameric torus. Our results are consistent with a model of local pH sensing at SctV, where pH-dependent dynamics of SctV cytoplasmic domain affect binding of gatekeeper complex. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
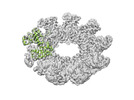
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11928.map.gz emd_11928.map.gz | 6.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11928-v30.xml emd-11928-v30.xml emd-11928.xml emd-11928.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11928_fsc.xml emd_11928_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11928.png emd_11928.png | 130 KB | ||
| Masks |  emd_11928_msk_1.map emd_11928_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11928.cif.gz emd-11928.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11928 http://ftp.pdbj.org/pub/emdb/structures/EMD-11928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11928 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11928 | HTTPS FTP |
-Related structure data
| Related structure data |  7awaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11928.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11928.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11928_msk_1.map emd_11928_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homo-nonameric ring complex of SsaV (SctV) type III secretion sys...
| Entire | Name: Homo-nonameric ring complex of SsaV (SctV) type III secretion system export apparatus protein. |
|---|---|
| Components |
|
-Supramolecule #1: Homo-nonameric ring complex of SsaV (SctV) type III secretion sys...
| Supramolecule | Name: Homo-nonameric ring complex of SsaV (SctV) type III secretion system export apparatus protein. type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Full length protein including transmembrane domain recombinantly expressed in E.coli C41 and extracted from membrane fraction with detergent DDM. Transmembrane domain present but not resolved. |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) / Strain: LT2 Salmonella enterica (bacteria) / Strain: LT2 |
| Molecular weight | Theoretical: 680 KDa |
-Macromolecule #1: Secretion system apparatus protein SsaV
| Macromolecule | Name: Secretion system apparatus protein SsaV / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 76.221359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRSWLGEGVR AQQWLSVCAG RQDMVLATVL LIAIVMMLLP LPTWMVDILI TINLMFSVIL LLIAIYLSDP LDLSVFPSLL LITTLYRLS LTISTSRLVL LQHNAGNIVD AFGKFVVGGN LTVGLVVFTI ITIVQFIVIT KGIERVAEVS ARFSLDGMPG K QMSIDGDL ...String: MRSWLGEGVR AQQWLSVCAG RQDMVLATVL LIAIVMMLLP LPTWMVDILI TINLMFSVIL LLIAIYLSDP LDLSVFPSLL LITTLYRLS LTISTSRLVL LQHNAGNIVD AFGKFVVGGN LTVGLVVFTI ITIVQFIVIT KGIERVAEVS ARFSLDGMPG K QMSIDGDL RAGVIDADHA RTLRQHVQQE SRFLGAMDGA MKFVKGDTIA GIIVVLVNII GGIIIAIVQY DMSMSEAVHT YS VLSIGDG LCGQIPSLLI SLSAGIIVTR VPGEKRQNLA TELSSQIARQ PQSLILTAVV LMLLALIPGF PFITLAFFSA LLA LPIILI RRKKSVVSAN GVEAPEKDSM VPGACPLILR LSPTLHSADL IRDIDAMRWF LFEDTGVPLP EVNIEVLPEP TEKL TVLLY QEPVFSLSIP AQADYLLIGA DASVVGDSQT LPNGMGQICW LTKDMAHKAQ GFGLDVFAGS QRISALLKCV LLRHM GEFI GVQETRYLMN AMEKNYSELV KELQRQLPIN KIAETLQRLV SERVSIRDLR LIFGTLIDWA PREKDVLMLT EYVRIA LRR HILRRLNPEG KPLPILRIGE GIENLVRESI RQTAMGTYTA LSSRHKTQIL QLIEQALKQS AKLFIVTSVD TRRFLRK IT EATLFDVPIL SWQELGEESL IQVVESIDLS EEELADNEEH HHHHH UniProtKB: Secretion system apparatus protein SsaV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: CONTINUOUS / Support film - #0 - Film thickness: 0.3 / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: HOLEY / Support film - #1 - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot 2.5s. | |||||||||||||||
| Details | Sample was monodisperse on a continuous carbon film. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 6729 / Average exposure time: 12.0 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: OTHER |
|---|---|
| Output model |  PDB-7awa: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)