[English] 日本語
 Yorodumi
Yorodumi- EMDB-22589: Cryo-EM structure of the nonameric EscV cytosolic domain from the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22589 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the nonameric EscV cytosolic domain from the type III secretion system | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Injectisome / Nonamer / Export Apparatus / Secretion / PROTEIN TRANSPORT | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |   | ||||||||||||
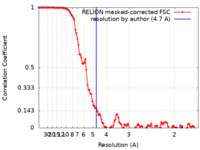
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | ||||||||||||
 Authors Authors | Majewski DD / Lyons BJE | ||||||||||||
| Funding support |  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2020 Journal: J Struct Biol / Year: 2020Title: Cryo-EM analysis of the SctV cytosolic domain from the enteropathogenic E. coli T3SS injectisome. Authors: Dorothy D Majewski / Bronwyn J E Lyons / Claire E Atkinson / Natalie C J Strynadka /  Abstract: The bacterial injectisome and flagella both rely on type III secretion systems for their assembly. The syringe-like injectisome creates a continuous channel between the bacterium and the host cell, ...The bacterial injectisome and flagella both rely on type III secretion systems for their assembly. The syringe-like injectisome creates a continuous channel between the bacterium and the host cell, through which signal-modulating effector proteins are secreted. The inner membrane pore protein SctV controls the hierarchy of substrate selection and may also be involved in energizing secretion. We present the 4.7 Å cryo-EM structure of the SctV cytosolic domain (SctV) from the enteropathogenic Escherichia coli injectisome. SctV forms a nonameric ring with primarily electrostatic interactions between its subunits. Molecular dynamics simulations show that monomeric SctV maintains a closed conformation, in contrast with previous studies on flagellar homologue FlhA. Comparison with substrate-bound homologues suggest that a conformational change would be required to accommodate binding partners. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22589.map.gz emd_22589.map.gz | 14.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22589-v30.xml emd-22589-v30.xml emd-22589.xml emd-22589.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22589_fsc.xml emd_22589_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22589.png emd_22589.png | 67.8 KB | ||
| Filedesc metadata |  emd-22589.cif.gz emd-22589.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22589 http://ftp.pdbj.org/pub/emdb/structures/EMD-22589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22589 | HTTPS FTP |
-Related structure data
| Related structure data |  7k08MC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22589.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22589.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.852 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EscV Cytosolic Nonamer Ring
| Entire | Name: EscV Cytosolic Nonamer Ring |
|---|---|
| Components |
|
-Supramolecule #1: EscV Cytosolic Nonamer Ring
| Supramolecule | Name: EscV Cytosolic Nonamer Ring / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Translocator EscV
| Macromolecule | Name: Translocator EscV / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: E2348/69 / EPEC |
| Molecular weight | Theoretical: 39.081633 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMADLSNS QNISPGAEPL ILNLSSNIYS SDITQQIEVM RWNFFEESGI PLPKIIVNPV KNNDSAIEFL LYQESIYKDT LIDDTVYFE AGHAEISFEF VQEKLSTNSI VYKTNKTNQQ LAHLTGMDVY ATTNDKITFL LKKLVLSNAK EFIGVQETRY L MDIMERKY ...String: GSHMADLSNS QNISPGAEPL ILNLSSNIYS SDITQQIEVM RWNFFEESGI PLPKIIVNPV KNNDSAIEFL LYQESIYKDT LIDDTVYFE AGHAEISFEF VQEKLSTNSI VYKTNKTNQQ LAHLTGMDVY ATTNDKITFL LKKLVLSNAK EFIGVQETRY L MDIMERKY NELVKELQRQ LGLSKIVDIL QRLVEENVSI RDLRTIFETL IFWSTKEKDV VILCEYVRIA LRRHILGRYS VS GTLLNVW LIGSDIENEL RESIRQTSSG SYLNISPERT EQIIGFLKNI MNPTGNGVIL TALDIRRYVK KMIEGSFPSV PVL SFQEVG NNIELKVLGT VNDFRA UniProtKB: Translocator EscV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)