+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
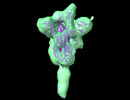
| Title | Prefusion spike of SARS-CoV-2 (Wuhan), 1-RBD-up conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | spike / SARS-CoV-2 / rbd up / receptor binding / VIRAL PROTEIN | ||||||||||||
| Biological species |  | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 27.75 Å | ||||||||||||
 Authors Authors | Calder LJ / Calcraft T / Hussain S / Harvey R / Rosenthal PB | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Electron cryotomography of SARS-CoV-2 virions reveals cylinder-shaped particles with a double layer RNP assembly. Authors: Lesley J Calder / Thomas Calcraft / Saira Hussain / Ruth Harvey / Peter B Rosenthal /  Abstract: SARS-CoV-2 is a lipid-enveloped Betacoronavirus and cause of the Covid-19 pandemic. To study the three-dimensional architecture of the virus, we perform electron cryotomography (cryo-ET) on SARS-Cov- ...SARS-CoV-2 is a lipid-enveloped Betacoronavirus and cause of the Covid-19 pandemic. To study the three-dimensional architecture of the virus, we perform electron cryotomography (cryo-ET) on SARS-Cov-2 virions and three variants revealing particles of regular cylindrical morphology. The ribonucleoprotein particles packaging the genome in the virion interior form a dense, double layer assembly with a cylindrical shape related to the overall particle morphology. This organisation suggests structural interactions important to virus assembly. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15185.map.gz emd_15185.map.gz | 566 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15185-v30.xml emd-15185-v30.xml emd-15185.xml emd-15185.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15185_fsc.xml emd_15185_fsc.xml | 3.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15185.png emd_15185.png | 50 KB | ||
| Masks |  emd_15185_msk_1.map emd_15185_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15185.cif.gz emd-15185.cif.gz | 4.6 KB | ||
| Others |  emd_15185_half_map_1.map.gz emd_15185_half_map_1.map.gz emd_15185_half_map_2.map.gz emd_15185_half_map_2.map.gz | 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15185 http://ftp.pdbj.org/pub/emdb/structures/EMD-15185 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15185 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15185 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15185.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15185.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.44 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15185_msk_1.map emd_15185_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15185_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15185_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / Details: Virus grown in Vero V1 cells and purified / NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Sci species strain: Wuhan / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum ER / Energy filter - Slit width: 20 eV |
| Details | Tilt series collected using dose symmetric scheme. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-4 / Average exposure time: 1.7 sec. / Average electron dose: 2.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)