-Search query
-Search result
Showing all 43 items for (author: arndt & m)

EMDB-19163: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

EMDB-19164: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.

EMDB-19165: 
Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

EMDB-19166: 
Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.

PDB-8rgz: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

PDB-8rh0: 
Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.

PDB-8rh1: 
Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules.

PDB-8rh2: 
Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules.
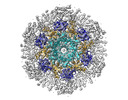
EMDB-29607: 
HIV-2 Gag Capsid from Immature Virus-like Particles

PDB-8fzc: 
HIV-2 Gag Capsid from Immature Virus-like Particles

EMDB-29606: 
CryoET of HIV-2 Immature Particles
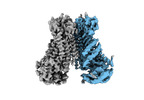
EMDB-15919: 
Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+

PDB-8b8q: 
Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+

EMDB-15913: 
Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin

EMDB-15914: 
Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin

EMDB-15916: 
Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed

EMDB-15917: 
Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed
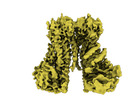
EMDB-15958: 
Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed

EMDB-15959: 
Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN

PDB-8b8g: 
Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin

PDB-8b8j: 
Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin

PDB-8b8k: 
Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed

PDB-8b8m: 
Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed

PDB-8bc0: 
Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed

PDB-8bc1: 
Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN

EMDB-11787: 
Cryo-EM structure of F-actin stabilized by cis-optoJASP-8

EMDB-11790: 
Cryo-EM structure of F-actin stabilized by trans-optoJASP-8

PDB-7ahn: 
Cryo-EM structure of F-actin stabilized by cis-optoJASP-8

PDB-7ahq: 
Cryo-EM structure of F-actin stabilized by trans-optoJASP-8

EMDB-4399: 
Cryo-EM structure of the RIP2 CARD filament

PDB-6ggs: 
Structure of RIP2 CARD filament

EMDB-3835: 
Cryo-EM structure of F-actin in complex with ADP

EMDB-3836: 
Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP

EMDB-3837: 
Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP-Pi

EMDB-3838: 
Cryo-EM structure of F-actin in complex with AppNHp (AMPPNP)

EMDB-3839: 
Cryo-EM structure of F-actin in complex with ADP-BeFx

EMDB-4259: 
Cryo-EM structure of F-actin in complex with ADP-Pi

PDB-5onv: 
Cryo-EM structure of F-actin in complex with ADP

PDB-5ooc: 
Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP

PDB-5ood: 
Cryo-EM structure of jasplakinolide-stabilized F-actin in complex with ADP-Pi

PDB-5ooe: 
Cryo-EM structure of F-actin in complex with AppNHp (AMPPNP)

PDB-5oof: 
Cryo-EM structure of F-actin in complex with ADP-BeFx

PDB-6fhl: 
Cryo-EM structure of F-actin in complex with ADP-Pi
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
