-Search query
-Search result
Showing 1 - 50 of 6,361 items for (author: ren & d)
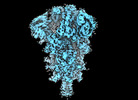
EMDB-50263: 
SARS-CoV-2 BA-2.87.1 Spike ectodomain
Method: single particle / : Ren J, Stuart DI, Duyvesteyn HME
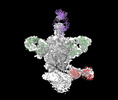
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.
Method: single particle / : Acharya P, Parsons R, Janowska K, Williams WB, Alam M, Haynes BF

EMDB-51031: 
Cryo-electron tomogram of AL59 amyloids interacting with collagen VI
Method: electron tomography / : Sicking K, Fernandez-Busnadiego R, Ricagno S

EMDB-51032: 
Cryo-electron tomogram of AL59 amyloids interacting with collagen VI
Method: electron tomography / : Sicking K, Fernandez-Busnadiego R, Ricagno S

EMDB-51033: 
Cryo-electron tomogram of AL59 amyloids interacting with collagen VI
Method: electron tomography / : Sicking K, Fernandez-Busnadiego R, Ricagno S

EMDB-51038: 
Cryo-electron tomogram of AL59 amyloids interacting with collagen VI
Method: electron tomography / : Sicking K, Fernandez-Busnadiego R, Ricagno S

EMDB-43746: 
Plasmodium falciparum 20S proteasome bound to an inhibitor
Method: single particle / : Han Y, Deng X, Ray S, Chen Z, Phillips M

PDB-8w2f: 
Plasmodium falciparum 20S proteasome bound to an inhibitor
Method: single particle / : Han Y, Deng X, Ray S, Chen Z, Phillips M
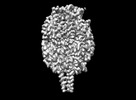
EMDB-43516: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-43517: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt2: 
cryo-EM structure of HMPV (MPV-2c)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt3: 
cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-37441: 
FCP tetramer in Chaetoceros gracilis
Method: single particle / : Feng Y, Li Z, Zhou C, Shen JR, Liu C, Wang W

EMDB-37442: 
FCP pentamer in Chaetoceros gracilis
Method: single particle / : Feng Y, Li Z, Zhou C, Liu C, Shen JR, Wang W

PDB-8wck: 
FCP tetramer in Chaetoceros gracilis
Method: single particle / : Feng Y, Li Z, Zhou C, Shen JR, Liu C, Wang W

PDB-8wcl: 
FCP pentamer in Chaetoceros gracilis
Method: single particle / : Feng Y, Li Z, Zhou C, Liu C, Shen JR, Wang W

EMDB-18973: 
Cryo-EM structure of Human SHMT1
Method: single particle / : Spizzichino S, Marabelli C, Bharadwaj A, Jakobi AJ, Chaves-Sanjuan A, Giardina G, Bolognesi M, Cutruzzola F

PDB-8r7h: 
Cryo-EM structure of Human SHMT1
Method: single particle / : Spizzichino S, Marabelli C, Bharadwaj A, Jakobi AJ, Chaves-Sanjuan A, Giardina G, Bolognesi M, Cutruzzola F

EMDB-18990: 
CryoEM map of tau PHF sarkosyl-extracted from a human AD patient (associated with in situ tomography)
Method: helical / : Wilkinson MW, Gilbert MAG, Fatima N, Jenkins J, O'Sullivan TJ, Schertel A, Halfon Y, Morrema THJ, Geibel M, Ranson NA, Radford SE, Hoozemans JJM, Frank RAW

EMDB-18689: 
Maps of Collagen VI half- and full-beads
Method: single particle / : Schulte T, Speranzini V, Chaves-Sanjuan A, Ricagno S

EMDB-50270: 
Cryo-EM structure of cardiac collagen-associated amyloid AL59
Method: helical / : Schulte T, Speranzini V, Chaves-Sanjuan A, Milazzo M, Ricagno S

PDB-9faa: 
Cryo-EM structure of cardiac collagen-associated amyloid AL59
Method: helical / : Schulte T, Speranzini V, Chaves-Sanjuan A, Milazzo M, Ricagno S

EMDB-44395: 
Full-length cross-linked Contactin 2 (CNTN2)
Method: single particle / : Liu JL, Fan SF, Ren GR, Rudenko GR

EMDB-44396: 
Cross-linked Contactin 2 Ig1-Ig6
Method: single particle / : Liu JL, Fan SF, Ren GR, Rudenko GR

EMDB-44397: 
Full-length cross-linked Contactin 2 (FN1 apart)
Method: single particle / : Liu JL, Fan SF, Ren GR, Rudenko GR

PDB-9ba4: 
Full-length cross-linked Contactin 2 (CNTN2)
Method: single particle / : Liu JL, Fan SF, Ren GR, Rudenko GR

PDB-9ba5: 
Cross-linked Contactin 2 Ig1-Ig6
Method: single particle / : Liu JL, Fan SF, Ren GR, Rudenko GR
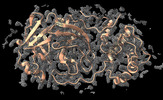
EMDB-43144: 
MicroED structure of SARS-CoV-2 main protease (MPro/3CLPro) with missing cone eliminated by suspended drop
Method: electron crystallography / : Bu G, Gillman C, Danelius E, Hattne J, Nannenga BL, Gonen T

PDB-8vd7: 
MicroED structure of SARS-CoV-2 main protease (MPro/3CLPro) with missing cone eliminated by suspended drop
Method: electron crystallography / : Bu G, Gillman C, Danelius E, Hattne J, Nannenga BL, Gonen T

EMDB-17691: 
60-meric complex of dihydrolipoamide acetyltransferase (E2) of the human pyruvate dehydrogenase complex (icosahedral symmetry)
Method: single particle / : Zdanowicz R, Afanasyev P, Boehringer D, Glockshuber R
EMDB-17694: 
60-meric complex of dihydrolipoamide acetyltransferase (E2) of the human pyruvate dehydrogenase complex (tetrahedral symmetry)
Method: single particle / : Zdanowicz R, Afanasyev P, Boehringer D, Glockshuber R

EMDB-18616: 
E2/E3BP core of the human pyruvate dehydrogenase complex (map 1; 3.4 A)
Method: single particle / : Zdanowicz R, Afanasyev P, Boehringer D, Glockshuber R

EMDB-18617: 
E2/E3BP core of the human pyruvate dehydrogenase complex (map 2; 3.7 A)
Method: single particle / : Zdanowicz R, Afanasyev P, Boehringer D, Glockshuber R

PDB-8piu: 
60-meric complex of dihydrolipoamide acetyltransferase (E2) of the human pyruvate dehydrogenase complex
Method: single particle / : Zdanowicz R, Afanasyev P, Boehringer D, Glockshuber R

EMDB-50148: 
Tau PHF subtomogram average relating to CS1 extended data Figure 9A
Method: subtomogram averaging / : Jenkins J

EMDB-50152: 
Tau PHF subtomogram average relating to CS2 Figure 3i-j.
Method: subtomogram averaging / : Jenkins J

EMDB-50153: 
Tau PHF subtomogram average relating to CS3 extended data Figure 9c
Method: subtomogram averaging / : Jenkins J

EMDB-50155: 
Tau PHF subtomogram average relating to CS4 extended data Figure 9d
Method: subtomogram averaging / : Jenkins J

EMDB-50156: 
Tau PHF subtomogram average relating to CS5 extended data Figure 9b
Method: subtomogram averaging / : Jenkins J

EMDB-50157: 
Tau PHF subtomogram average relating to CS6 extended data Figure 9e
Method: subtomogram averaging / : Jenkins J

EMDB-50159: 
Tau PHF subtomogram average relating to CS7 extended data Figure 9f
Method: subtomogram averaging / : Jenkins J

EMDB-50160: 
Tau PHF subtomogram average relating to LOL1_PHF Figure 4g-h
Method: subtomogram averaging / : Jenkins J

EMDB-50161: 
Tau SF subtomogram average relating to LOL1_SF Figure 4g-h
Method: subtomogram averaging / : Jenkins J

EMDB-50162: 
Tau SF subtomogram average relating to LOL2_SF Figure 4i-j
Method: subtomogram averaging / : Jenkins J

EMDB-43296: 
Constituent EM map: Focused refinement S100A1 of mouse RyR1 in complex with S100A1 (EGTA-only dataset)
Method: single particle / : Weninger G, Marks AR

EMDB-50580: 
SOLIST cryo-tomogram of native left ventricle mouse heart muscle #1
Method: electron tomography / : Erdmann PS, Nguyen HTD, Perone G, Klena N, Vazzana R, Kaluthantrige Don F, Silva M, Sorrentino S, Swuec P, Leroux F, Kalebic N, Coscia F

EMDB-50582: 
SOLIST native mouse heart muscle tomogram #2
Method: electron tomography / : Erdmann PS, Nguyen HTD, Perone G, Klena N, Vazzana R, Kaluthantrige Don F, Silva M, Sorrentino S, Swuec P, Leroux F, Kalebic N, Coscia F

EMDB-37546: 
Spike Trimer of BA.2.86 in complex with one hACE2
Method: single particle / : Yue C, Liu P

EMDB-37548: 
Spike Trimer of BA.2.86 in complex with two hACE2s
Method: single particle / : Yue C, Liu P

EMDB-37549: 
Spike Trimer of BA.2.86 with three RBDs down
Method: single particle / : Yue C, Liu P
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
