+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 BA-2.87.1 Spike ectodomain | |||||||||
 Map data Map data | BA2-87.1 cryoEM map C3 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral protein / immune system / SARS-CoV-2 / RBD / Spike / glycoprotein / BA.2.87.1 / receptor / coronavirus-2 / N-terminal domain / Supersite | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...virion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Ren J / Stuart DI / Duyvesteyn HME | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike. Authors: Helen M E Duyvesteyn / Aiste Dijokaite-Guraliuc / Chang Liu / Piyada Supasa / Barbara Kronsteiner / Katie Jeffery / Lizzie Stafford / Paul Klenerman / Susanna J Dunachie / / Juthathip ...Authors: Helen M E Duyvesteyn / Aiste Dijokaite-Guraliuc / Chang Liu / Piyada Supasa / Barbara Kronsteiner / Katie Jeffery / Lizzie Stafford / Paul Klenerman / Susanna J Dunachie / / Juthathip Mongkolsapaya / Elizabeth E Fry / Jingshan Ren / David I Stuart / Gavin R Screaton /   Abstract: BA.2.87.1 represents a major shift in the BA.2 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and is unusual in having two lengthy deletions of polypeptide in the spike (S) ...BA.2.87.1 represents a major shift in the BA.2 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and is unusual in having two lengthy deletions of polypeptide in the spike (S) protein, one of which removes a beta-strand. Here we investigate its neutralization by a variety of sera from infected and vaccinated individuals and determine its spike (S) ectodomain structure. The BA.2.87.1 receptor binding domain (RBD) is structurally conserved and the RBDs are tightly packed in an "all-down" conformation with a small rotation relative to the trimer axis as compared to the closest previously observed conformation. The N-terminal domain (NTD) maintains a remarkably similar structure overall; however, the rearrangements resulting from the deletions essentially destroy the so-called supersite epitope and eliminate one glycan site, while a mutation creates an additional glycan site, effectively shielding another NTD epitope. BA.2.87.1 is relatively easily neutralized but acquisition of additional mutations in the RBD could increase antibody escape allowing it to become a dominant sub-lineage. #1:  Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2019 Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2019Title: Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix Authors: Liebschner D / Afonine PV #2:  Journal: Nature Methods / Year: 2017 Journal: Nature Methods / Year: 2017Title: cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination Authors: Punjani A / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50263.map.gz emd_50263.map.gz | 151.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50263-v30.xml emd-50263-v30.xml emd-50263.xml emd-50263.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50263.png emd_50263.png | 85.5 KB | ||
| Filedesc metadata |  emd-50263.cif.gz emd-50263.cif.gz | 7.6 KB | ||
| Others |  emd_50263_half_map_1.map.gz emd_50263_half_map_1.map.gz emd_50263_half_map_2.map.gz emd_50263_half_map_2.map.gz | 147.6 MB 151 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50263 http://ftp.pdbj.org/pub/emdb/structures/EMD-50263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50263 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50263 | HTTPS FTP |
-Related structure data
| Related structure data |  9f9yMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50263.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50263.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BA2-87.1 cryoEM map C3 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7303 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: BA2-87.1 cryoEM half map C3 symmetry
| File | emd_50263_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BA2-87.1 cryoEM half map C3 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
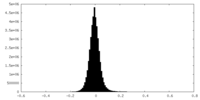
| Density Histograms |
-Half map: BA2-87.1 cryoEM half map C3 symmetry
| File | emd_50263_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BA2-87.1 cryoEM half map C3 symmetry | ||||||||||||
| Projections & Slices |
| ||||||||||||
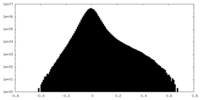
| Density Histograms |
- Sample components
Sample components
-Entire : BA-2.87.1 variant SARS-CoV2 S protein
| Entire | Name: BA-2.87.1 variant SARS-CoV2 S protein |
|---|---|
| Components |
|
-Supramolecule #1: BA-2.87.1 variant SARS-CoV2 S protein
| Supramolecule | Name: BA-2.87.1 variant SARS-CoV2 S protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Spike protein recombinantly expressed using sequence of human-derived BA-2.87.1 variant of SARS-CoV2. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 139.502484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQSYTNSF TRGVYYPDKV FRSSVLHSTQ DLFLPFFSNV TWFHAISGTN DTKRFDNPVL PFNDGVYFAS TEKFNIIRG WIFGTTLDSK TQSLLIVNNA TNAVIKVCEF QFKNNKSLME SEFRVYSSAN NCTFEYVSQP FLMDLEGKQG N FKNLSEFV ...String: MFVFLVLLPL VSSQSYTNSF TRGVYYPDKV FRSSVLHSTQ DLFLPFFSNV TWFHAISGTN DTKRFDNPVL PFNDGVYFAS TEKFNIIRG WIFGTTLDSK TQSLLIVNNA TNAVIKVCEF QFKNNKSLME SEFRVYSSAN NCTFEYVSQP FLMDLEGKQG N FKNLSEFV FKNIDGYFKI YSKHTPINLG RGLPQGFSAL EPLVDLPIGI NITRFQTLLA LHRSYLTPGD SSSGWTAGAA AY YVGYLQP RTFLLKYNEN GTITDAVDCA LDPLSETKCT LKSFTVEKGI YQTSNFRVQP TESIVRFPNI TNLCPFDEVF NAT RFASVY AWNRKRISNC VADYSVLYNF APFFAFKCYG VSPTKLNDLC FTNVYADSFV IRGNEVSQIA PGQTGTIADY NYKL PDDFT GCVIAWNSNK LDSNGGGNYN YMYRLFRKSK LKPFERDIST EIYQAGNTPC KGVAGFNCYF PLQSYGFRPT YGVGH QPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLTGT GVLTESNKKF LPFQQFGRDI ADTTDAVRDP QTLEIL DIT PCSFGGVSVI TPGTNTSNQV AVLYQGVNCT EVSVAIHADQ LTPTWRVYST GSNGFQTRAG CLIGAEYVNN SYECDIP IG AGICASYQTQ TRSHGSASSV ASQPIIAYTM SLGAENSVAY SNNSIAIPTN FTISVTTEIL PVSMTKTSVD CTMYICGD S TECSNLLLQY GSFCTQLKRA LTGIAVEQDK NTQEVFAQVK QIYKIPPIKH FGGFNFSQIL PDPSKPSKRS FIEDLLFNK VTLADAGFIK QYGDCLGDIA ARDLICAQKF NGLTVLPPLL TDEMIAQYTS ALLAGTITSG WTFGAGAALQ IPFAMQMAYR FNGIGVTQN VLYENQKLIA NQFNSAIGKI QGSLSSTASA LGKLQDVVNH NAQALNTLVK QLSSKFGAIS SVLNDILSRL D PPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QSAPHGVVFL HV TYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRNFYEP QIITTDNTFV SGNCDVVIGI VNNTVYDPLQ PEL DSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLNESLIDLQ ELGKYEQGSG YIPEAPRDGQ AYVR KDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHGSA WSHPQFEKGG GSGGGSGGSA WSHPQFEK UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 39 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
| Details | BA.2.87.1 Spike |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 9818 / Average electron dose: 40.0 e/Å2 / Details: Images were collected in EER format. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)