+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Maps of Collagen VI half- and full-beads | |||||||||
 Map data Map data | full-bead map, with one of the half-beads not in focus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Collagen / Extracellular matrix / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to polyamine macromolecule / response to bleomycin / regulation of collagen fibril organization / muscle cell apoptotic process / collagen type VI trimer / multicellular organismal locomotion / muscle system process / apoptotic nuclear changes / limb joint morphogenesis / caveola assembly ...response to polyamine macromolecule / response to bleomycin / regulation of collagen fibril organization / muscle cell apoptotic process / collagen type VI trimer / multicellular organismal locomotion / muscle system process / apoptotic nuclear changes / limb joint morphogenesis / caveola assembly / mitochondrial transmembrane transport / reduction of food intake in response to dietary excess / skeletal muscle tissue growth / fat cell proliferation / extracellular matrix assembly / response to decreased oxygen levels / Collagen chain trimerization / platelet-derived growth factor binding / extracellular matrix structural constituent conferring tensile strength / response to peptide / tissue remodeling / skeletal muscle fiber differentiation / lung epithelial cell differentiation / basement membrane organization / sensory perception of mechanical stimulus / Collagen biosynthesis and modifying enzymes / collagen metabolic process / Signaling by PDGF / energy reserve metabolic process / mitochondrial depolarization / 2-oxoglutarate metabolic process / respiratory system process / skeletal muscle tissue regeneration / myelination in peripheral nervous system / NCAM1 interactions / cartilage development / collagen fibril organization / Assembly of collagen fibrils and other multimeric structures / lung alveolus development / response to pain / muscle organ development / response to muscle activity / regulation of cell size / bone mineralization / intracellular vesicle / myofibril / endodermal cell differentiation / Collagen degradation / uterus development / lung morphogenesis / transmission of nerve impulse / basement membrane / homeostasis of number of cells / hair follicle development / canonical Wnt signaling pathway / ECM proteoglycans / adipose tissue development / Integrin cell surface interactions / response to mechanical stimulus / single fertilization / response to glucose / response to UV / collagen binding / skeletal muscle fiber development / tricarboxylic acid cycle / insulin-like growth factor receptor signaling pathway / reactive oxygen species metabolic process / response to reactive oxygen species / glycolytic process / sarcoplasmic reticulum / mitochondrion organization / phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein tetramerization / cellular response to amino acid stimulus / serine-type endopeptidase inhibitor activity / circadian rhythm / sarcolemma / bone development / : / autophagy / response to wounding / response to toxic substance / extracellular matrix / cell morphogenesis / osteoblast differentiation / insulin receptor signaling pathway / extracellular vesicle / heart development / neuron apoptotic process / gene expression / response to lipopolysaccharide / cell adhesion / endoplasmic reticulum lumen / response to xenobiotic stimulus / inflammatory response / lysosomal membrane / protein-containing complex / mitochondrion / extracellular space / extracellular exosome Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
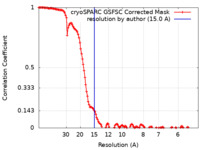
| Method | single particle reconstruction / cryo EM / Resolution: 15.0 Å | |||||||||
 Authors Authors | Schulte T / Speranzini V / Chaves-Sanjuan A / Ricagno S | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis. Authors: Tim Schulte / Antonio Chaves-Sanjuan / Valentina Speranzini / Kevin Sicking / Melissa Milazzo / Giulia Mazzini / Paola Rognoni / Serena Caminito / Paolo Milani / Chiara Marabelli / ...Authors: Tim Schulte / Antonio Chaves-Sanjuan / Valentina Speranzini / Kevin Sicking / Melissa Milazzo / Giulia Mazzini / Paola Rognoni / Serena Caminito / Paolo Milani / Chiara Marabelli / Alessandro Corbelli / Luisa Diomede / Fabio Fiordaliso / Luigi Anastasia / Carlo Pappone / Giampaolo Merlini / Martino Bolognesi / Mario Nuvolone / Rubén Fernández-Busnadiego / Giovanni Palladini / Stefano Ricagno /     Abstract: Systemic light chain (LC) amyloidosis (AL) is a disease where organs are damaged by an overload of a misfolded patient-specific antibody-derived LC, secreted by an abnormal B cell clone. The high LC ...Systemic light chain (LC) amyloidosis (AL) is a disease where organs are damaged by an overload of a misfolded patient-specific antibody-derived LC, secreted by an abnormal B cell clone. The high LC concentration in the blood leads to amyloid deposition at organ sites. Indeed, cryogenic electron microscopy (cryo-EM) has revealed unique amyloid folds for heart-derived fibrils taken from different patients. Here, we present the cryo-EM structure of heart-derived AL amyloid (AL59) from another patient with severe cardiac involvement. The double-layered structure displays a u-shaped core that is closed by a β-arc lid and extended by a straight tail. Noteworthy, the fibril harbours an extended constant domain fragment, thus ruling out the variable domain as sole amyloid building block. Surprisingly, the fibrils were abundantly concatenated with a proteinaceous polymer, here identified as collagen VI (COLVI) by immuno-electron microscopy (IEM) and mass-spectrometry. Cryogenic electron tomography (cryo-ET) showed how COLVI wraps around the amyloid forming a helical superstructure, likely stabilizing and protecting the fibrils from clearance. Thus, here we report structural evidence of interactions between amyloid and collagen, potentially signifying a distinct pathophysiological mechanism of amyloid deposits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18689.map.gz emd_18689.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18689-v30.xml emd-18689-v30.xml emd-18689.xml emd-18689.xml | 28.5 KB 28.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18689_fsc.xml emd_18689_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_18689.png emd_18689.png | 33 KB | ||
| Masks |  emd_18689_msk_1.map emd_18689_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18689.cif.gz emd-18689.cif.gz | 8.8 KB | ||
| Others |  emd_18689_additional_1.map.gz emd_18689_additional_1.map.gz emd_18689_additional_2.map.gz emd_18689_additional_2.map.gz emd_18689_half_map_1.map.gz emd_18689_half_map_1.map.gz emd_18689_half_map_2.map.gz emd_18689_half_map_2.map.gz | 111.9 MB 113.8 MB 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18689 http://ftp.pdbj.org/pub/emdb/structures/EMD-18689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18689 | HTTPS FTP |
-Related structure data
| Related structure data |  9faaC  9facC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18689.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18689.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full-bead map, with one of the half-beads not in focus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.607 Å | ||||||||||||||||||||||||||||||||||||
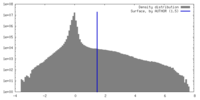
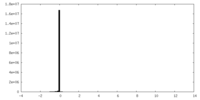
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18689_msk_1.map emd_18689_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
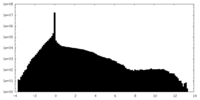
| Density Histograms |
-Additional map: #1
| File | emd_18689_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
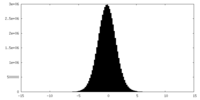
| Density Histograms |
-Additional map: composite map obtained by fitting the focused half-bead...
| File | emd_18689_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map obtained by fitting the focused half-bead map into the unfocused half-bead of the full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
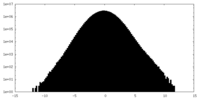
| Density Histograms |
-Half map: half-map A associated with main map
| File | emd_18689_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map A associated with main map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map B associated with main map
| File | emd_18689_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map B associated with main map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Collagen VI bead
| Entire | Name: Collagen VI bead |
|---|---|
| Components |
|
-Supramolecule #1: Collagen VI bead
| Supramolecule | Name: Collagen VI bead / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Collagen VI co-extracted with light chain amyloid AL59 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: HEART Homo sapiens (human) / Organ: HEART |
-Macromolecule #1: CO6A1_HUMAN; Collagen alpha-1(VI) chain
| Macromolecule | Name: CO6A1_HUMAN; Collagen alpha-1(VI) chain / type: protein_or_peptide / ID: 1 / Details: CO6A1_HUMAN; Collagen alpha-1(VI) chain; P12109 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: HEART Homo sapiens (human) / Organ: HEART |
| Sequence | String: MRAARALLPL LLQACWTAAQ DEPETPRAVA FQDCPVDLFF VLDTSESVAL RLKPYGALVD KVKSFTKRFI DNLRDRYYRC DRNLVWNAGA LHYSDEVEII QGLTRMPGGR DALKSSVDAV KYFGKGTYTD CAIKKGLEQL LVGGSHLKEN KYLIVVTDGH PLEGYKEPCG ...String: MRAARALLPL LLQACWTAAQ DEPETPRAVA FQDCPVDLFF VLDTSESVAL RLKPYGALVD KVKSFTKRFI DNLRDRYYRC DRNLVWNAGA LHYSDEVEII QGLTRMPGGR DALKSSVDAV KYFGKGTYTD CAIKKGLEQL LVGGSHLKEN KYLIVVTDGH PLEGYKEPCG GLEDAVNEAK HLGVKVFSVA ITPDHLEPRL SIIATDHTYR RNFTAADWGQ SRDAEEAISQ TIDTIVDMIK NNVEQVCCSF ECQPARGPPG LRGDPGFEGE RGKPGLPGEK GEAGDPGRPG DLGPVGYQGM KGEKGSRGE KGSRGPKGYK GEKGKRGIDG VDGVKGEMGY PGLPGCKGSP GFDGIQGPPG PKGDPGAFGL KGEKGEPGAD GEAGRPGSSG PSGDEGQPGE PGPPGEKGEA GDEGNPGPDG APGERGGPGE RGPRGTPGTR GPRGDPGEAG PQGDQGREGP VGVPGDPGEA GPIGPKGYRG DEGPPGSEGA RGAPGPAGPP GDPGLMGERG EDGPAGNGTE GFPGFPGYPG NRGAPGINGT KGYPGLKGDE GEAGDPGDDN NDIAPRGVKG AKGYRGPEGP QGPPGHQGPP GPDECEILDI IMKMCSCCEC KCGPIDLLFV LDSSESIGLQ NFEIAKDFVV KVIDRLSRDE LVKFEPGQSY AGVVQYSHSQ MQEHVSLRSP SIRNVQELKE AIKSLQWMAG GTFTGEALQY TRDQLLPPSP NNRIALVITD GRSDTQRDTT PLNVLCSPGI QVVSVGIKDV FDFIPGSDQL NVISCQGLAP SQGRPGLSLV KENYAELLED AFLKNVTAQI CIDKKCPDYT CPITFSSPAD ITILLDGSAS VGSHNFDTTK RFAKRLAERF LTAGRTDPAH DVRVAVVQYS GTGQQRPERA SLQFLQNYTA LASAVDAMDF INDATDVNDA LGYVTRFYRE ASSGAAKKRL LLFSDGNSQG ATPAAIEKAV Q EAQRAGIE IFVVVVGRQV NEPHIRVLVT GKTAEYDVAY GESHLFRVPS YQALLRGVFH QTVSRKVALG UniProtKB: Collagen alpha-1(VI) chain |
-Macromolecule #2: CO6A2_HUMAN; Collagen alpha-2(VI) chain
| Macromolecule | Name: CO6A2_HUMAN; Collagen alpha-2(VI) chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: HEART Homo sapiens (human) / Organ: HEART |
| Sequence | String: MLQGTCSVLL LWGILGAIQA QQQEVISPDT TERNNNCPEK TDCPIHVYFV LDTSESVTMQ SPTDILLFHM KQFVPQFISQ LQNEFYLDQV ALSWRYGGLH FSDQVEVFSP PGSDRASFIK NLQGISSFRR GTFTDCALAN MTEQIRQDRS KGTVHFAVVI TDGHVTGSPC ...String: MLQGTCSVLL LWGILGAIQA QQQEVISPDT TERNNNCPEK TDCPIHVYFV LDTSESVTMQ SPTDILLFHM KQFVPQFISQ LQNEFYLDQV ALSWRYGGLH FSDQVEVFSP PGSDRASFIK NLQGISSFRR GTFTDCALAN MTEQIRQDRS KGTVHFAVVI TDGHVTGSPC GGIKLQAERA REEGIRLFAV APNQNLKEQG LRDIASTPHE LYRNDYATML PDSTEIDQDT INRIIKVMKH EAYGECYKVS CLEIPGPSGP KGYRGQKGAK GNMGEPGEPG QKGRQGDPGI EGPIGFPGPK GVPGFKGEKG EFGADGRKGA PGLAGKNGTD GQKGKLGRIG PPGCKGDPGN RGPDGYPGEA GSPGERGDQG GKGDPGRPGR RGPPGEIGAK GSKGYQGNSG APGSPGVKGA KGGPGPRGPK GEPGRRGDPG TKGSPGSDGP KGEKGDPGPE GPRGLAGEVG NKGAKGDRGL PGPRGPQGAL GEPGKQGSRG DPGDAGPRGD SGQPGPKGDP GRPGFSYPGP RGAPGEKGEP GPRGPEGGRG DFGLKGEPGR KGEKGEPADP GPPGEPGPRG PRGVPGPEGE PGPPGDPGLT ECDVMTYVRE TCGCCDCEKR CGALDVVFVI DSSESIGYTN FTLEKNFVIN VVNRLGAIAK DPKSETGTRV GVVQYSHEGT FEAIQLDDER IDSLSSFKEA VKNLEWIAGG TWTPSALKFA YDRLIKESRR QKTRVFAVVI TDGRHDPRDD DLNLRALCDR DVTVTAIGIG DMFHEKHESE NLYSIACDKP QQVRNMTLFS DLVAEKFIDD MEDVLCPDPQ IVCPDLPCQT ELSVAQCTQR PVDIVFLLDG SERLGEQNFH KARRFVEQVA RRLTLARRDD DPLNARVALL QFGGPGEQQV AFPLSHNLTA IHEALETTQY LNSFSHVGAG VVHAINAIVR SPRGGARRHA ELSFVFLTDG VTGNDSLHES AHSMRKQNVV PTVLALGSDV DMDVLTTLSL GDRAAVFHEK DYDSLAQPGF FDRFIRWIC UniProtKB: Collagen alpha-2(VI) chain |
-Macromolecule #3: CO6A3_HUMAN; Collagen alpha-3(VI) chain
| Macromolecule | Name: CO6A3_HUMAN; Collagen alpha-3(VI) chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: HEART Homo sapiens (human) / Organ: HEART |
| Sequence | String: MRKHRHLPLV AVFCLFLSGF PTTHAQQQQA DVKNGAAADI IFLVDSSWTI GEEHFQLVRE FLYDVVKSLA VGENDFHFAL VQFNGNPHTE FLLNTYRTKQ EVLSHISNMS YIGGTNQTGK GLEYIMQSHL TKAAGSRAGD GVPQVIVVLT DGHSKDGLAL PSAELKSADV ...String: MRKHRHLPLV AVFCLFLSGF PTTHAQQQQA DVKNGAAADI IFLVDSSWTI GEEHFQLVRE FLYDVVKSLA VGENDFHFAL VQFNGNPHTE FLLNTYRTKQ EVLSHISNMS YIGGTNQTGK GLEYIMQSHL TKAAGSRAGD GVPQVIVVLT DGHSKDGLAL PSAELKSADV NVFAIGVEDA DEGALKEIAS EPLNMHMFNL ENFTSLHDIV GNLVSCVHSS VSPERAGDTE TLKDITAQDS ADIIFLIDGS NNTGSVNFAV ILDFLVNLLE KLPIGTQQIR VGVVQFSDEP RTMFSLDTYS TKAQVLGAVK ALGFAGGELA NIGLALDFVV ENHFTRAGGS RVEEGVPQVL VLISAGPSSD EIRYGVVALK QASVFSFGLG AQAASRAELQ HIATDDNLVF TVPEFRSFGD LQEKLLPYIV GVAQRHIVLK PPTIVTQVIE VNKRDIVFLV DGSSALGLAN FNAIRDFIAK VIQRLEIGQD LIQVAVAQYA DTVRPEFYFN THPTKREVIT AVRKMKPLDG SALYTGSALD FVRNNLFTSS AGYRAAEGIP KLLVLITGGK SLDEISQPAQ ELKRSSIMAF AIGNKGADQA ELEEIAFDSS LVFIPAEFRA APLQGMLPGL LAPLRTLSGT PEVHSNKRDI IFLLDGSANV GKTNFPYVRD FVMNLVNSLD IGNDNIRVGL VQFSDTPVTE FSLNTYQTKS DILGHLRQLQ LQGGSGLNTG SALSYVYANH FTEAGGSRIR EHVPQLLLLL TAGQSEDSYL QAANALTRAG ILTFCVGASQ ANKAELEQIA FNPSLVYLMD DFSSLPALPQ QLIQPLTTYV SGGVEEVPLA QPESKRDILF LFDGSANLVG QFPVVRDFLY KIIDELNVKP EGTRIAVAQY SDDVKVESRF DEHQSKPEIL NLVKRMKIKT GKALNLGYAL DYAQRYIFVK SAGSRIEDGV LQFLVLLVAG RSSDRVDGPA SNLKQSGVVP FIFQAKNADP AELEQIVLSP AFILAAESLP KIGDLHPQIV NLLKSVHNGA PAPVSGEKDV VFLLDGSEGV RSGFPLLKEF VQRVVESLDV GQDRVRVAVV QYSDRTRPEF YLNSYMNKQD VVNAVRQLTL LGGPTPNTGA ALEFVLRNIL VSSAGSRITE GVPQLLIVLT ADRSGDDVRN PSVVVKRGGA VPIGIGIGNA DITEMQTISF IPDFAVAIPT FRQLGTVQQV ISERVTQLTR EELSRLQPVL QPLPSPGVGG KRDVVFLIDG SQSAGPEFQY VRTLIERLVD YLDVGFDTTR VAVIQFSDDP KVEFLLNAHS SKDEVQNAVQ RLRPKGGRQI NVGNALEYVS RNIFKRPLGS RIEEGVPQFL VLISSGKSDD EVDDPAVELK QFGVAPFTIA RNADQEELVK ISLSPEYVFS VSTFRELPSL EQKLLTPITT LTSEQIQKLL ASTRYPPPAV ESDAADIVFL IDSSEGVRPD GFAHIRDFVS RIVRRLNIGP SKVRVGVVQF SNDVFPEFYL KTYRSQAPVL DAIRRLRLRG GSPLNTGKAL EFVARNLFVK SAGSRIEDGV PQHLVLVLGG KSQDDVSRFA QVIRSSGIVS LGVGDRNIDR TELQTITNDP RLVFTVREFR ELPNIEERIM NSFGPSAATP APPGVDTPPP SRPEKKKADI VFLLDGSINF RRDSFQEVLR FVSEIVDTVY EDGDSIQVGL VQYNSDPTDE FFLKDFSTKR QIIDAINKVV YKGGRHANTK VGLEHLRVNH FVPEAGSRLD QRVPQIAFVI TGGKSVEDAQ DVSLALTQRG VKVFAVGVRN IDSEEVGKIA SNSATAFRVG NVQELSELSE QVLETLHDAM HETLCPGVTD AAKACNLDVI LGFDGSRDQN VFVAQKGFES KVDAILNRIS QMHRVSCSGG RSPTVRVSVV ANTPSGPVEA FDFDEYQPEM LEKFRNMRSQ HPYVLTEDTL KVYLNKFRQS SPDSVKVVIH FTDGADGDLA DLHRASENLR QEGVRALILV GLERVVNLER LMHLEFGRGF MYDRPLRLNL LDLDYELAEQ LDNIAEKACC GVPCKCSGQR GDRGPIGSIG PKGIPGEDGY RGYPGDEGGP GERGPPGVNG TQGFQGCPGQ RGVKGSRGFP GEKGEVGEIG LDGLDGEDGD KGLPGSSGEK GNPGRRGDKG PRGEKGERGD VGIRGDPGNP GQDSQERGPK GETGDLGPMG VPGRDGVPGG PGETGKNGGF GRRGPPGAKG NKGGPGQPGF EGEQGTRGAQ GPAGPAGPPG LIGEQGISGP RGSGGAAGAP GERGRTGPLG RKGEPGEPGP KGGIGNRGPR GETGDDGRDG VGSEGRRGKK GERGFPGYPG PKGNPGEPGL NGTTGPKGIR GRRGNSGPPG IVGQKGDPGY PGPAGPKGNR GDSIDQCALI QSIKDKCPCC YGPLECPVFP TELAFALDTS EGVNQDTFGR MRDVVLSIVN DLTIAESNCP RGARVAVVTY NNEVTTEIRF ADSKRKSVLL DKIKNLQVAL TSKQQSLETA MSFVARNTFK RVRNGFLMRK VAVFFSNTPT RASPQLREAV LKLSDAGITP LFLTRQEDRQ LINALQINNT AVGHALVLPA GRDLTDFLEN VLTCHVCLDI CNIDPSCGFG SWRPSFRDRR AAGSDVDIDM AFILDSAETT TLFQFNEMKK YIAYLVRQLD MSPDPKASQH FARVAVVQHA PSESVDNASM PPVKVEFSLT DYGSKEKLVD FLSRGMTQLQ GTRALGSAIE YTIENVFESA PNPRDLKIVV LMLTGEVPEQ QLEEAQRVIL QAKCKGYFFV VLGIGRKVNI KEVYTFASEP NDVFFKLVDK STELNEEPLM RFGRLLPSFV SSENAFYLSP DIRKQCDWFQ GDQPTKNLVK FGHKQVNVPN NVTSSPTSNP VTTTKPVTTT KPVTTTTKPV TTTTKPVTII NQPSVKPAAA KPAPAKPVAA KPVATKMATV RPPVAVKPAT AAKPVAAKPA AVRPPAAAAA KPVATKPEVP RPQAAKPAAT KPATTKPMVK MSREVQVFEI TENSAKLHWE RAEPPGPYFY DLTVTSAHDQ SLVLKQNLTV TDRVIGGLLA GQTYHVAVVC YLRSQVRATY HGSFSTKKSQ PPPPQPARSA SSSTINLMVS TEPLALTETD ICKLPKDEGT CRDFILKWYY DPNTKSCARF WYGGCGGNEN KFGSQKECEK VCAPVLAKPG VISVMGT UniProtKB: Collagen alpha-3(VI) chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 1 / Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 2049 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 120000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 2 / Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 2556 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 73000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)