+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HMPV (MPV-2cREKR) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Prefusion / HMPV / Cryo-EM / VIRUS / VIRAL PROTEIN | |||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | |||||||||
| Biological species |  Human metapneumovirus Human metapneumovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Yu X / Langedijk JPM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer. Authors: Mark J G Bakkers / Tina Ritschel / Machteld Tiemessen / Jacobus Dijkman / Angelo A Zuffianò / Xiaodi Yu / Daan van Overveld / Lam Le / Richard Voorzaat / Marlies M van Haaren / Martijn de ...Authors: Mark J G Bakkers / Tina Ritschel / Machteld Tiemessen / Jacobus Dijkman / Angelo A Zuffianò / Xiaodi Yu / Daan van Overveld / Lam Le / Richard Voorzaat / Marlies M van Haaren / Martijn de Man / Sem Tamara / Leslie van der Fits / Roland Zahn / Jarek Juraszek / Johannes P M Langedijk /   Abstract: The prefusion conformation of human metapneumovirus fusion protein (hMPV Pre-F) is critical for eliciting the most potent neutralizing antibodies and is the preferred immunogen for an efficacious ...The prefusion conformation of human metapneumovirus fusion protein (hMPV Pre-F) is critical for eliciting the most potent neutralizing antibodies and is the preferred immunogen for an efficacious vaccine against hMPV respiratory infections. Here we show that an additional cleavage event in the F protein allows closure and correct folding of the trimer. We therefore engineered the F protein to undergo double cleavage, which enabled screening for Pre-F stabilizing substitutions at the natively folded protomer interfaces. To identify these substitutions, we developed an AI convolutional classifier that successfully predicts complex polar interactions often overlooked by physics-based methods and visual inspection. The combination of additional processing, stabilization of interface regions and stabilization of the membrane-proximal stem, resulted in a Pre-F protein vaccine candidate without the need for a heterologous trimerization domain that exhibited high expression yields and thermostability. Cryo-EM analysis shows the complete ectodomain structure, including the stem, and a specific interaction of the newly identified cleaved C-terminus with the adjacent protomer. Importantly, the protein induces high and cross-neutralizing antibody responses resulting in near complete protection against hMPV challenge in cotton rats, making the highly stable, double-cleaved hMPV Pre-F trimer an attractive vaccine candidate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43517.map.gz emd_43517.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43517-v30.xml emd-43517-v30.xml emd-43517.xml emd-43517.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43517.png emd_43517.png | 39.4 KB | ||
| Filedesc metadata |  emd-43517.cif.gz emd-43517.cif.gz | 6.5 KB | ||
| Others |  emd_43517_half_map_1.map.gz emd_43517_half_map_1.map.gz emd_43517_half_map_2.map.gz emd_43517_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43517 http://ftp.pdbj.org/pub/emdb/structures/EMD-43517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43517 | HTTPS FTP |
-Related structure data
| Related structure data |  8vt3MC  8vt2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43517.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43517.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
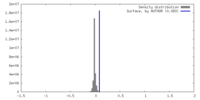
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43517_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
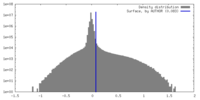
| Density Histograms |
-Half map: #1
| File | emd_43517_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
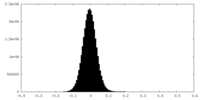
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion of HMPV (MPV-2cREKR) trimer complex
| Entire | Name: Prefusion of HMPV (MPV-2cREKR) trimer complex |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion of HMPV (MPV-2cREKR) trimer complex
| Supramolecule | Name: Prefusion of HMPV (MPV-2cREKR) trimer complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
-Macromolecule #1: Prefusion of HMPV (MPV-2cREKR), N-fragment
| Macromolecule | Name: Prefusion of HMPV (MPV-2cREKR), N-fragment / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 8.296239 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLKESYLEES CSTITEGYLS VLRTGWYTNV FTLEVGDVEN LTCSDGPSLI KTELDLTKSA LRELKTVSAD QLARE UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: Prefusion of HMPV (MPV-2cREKR), C-fragment
| Macromolecule | Name: Prefusion of HMPV (MPV-2cREKR), C-fragment / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 41.865828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FVLGAIALGR ATAAAVTAGV AIAKTIRLES EVTAIKNALK TTNEAVSTLG NGVRVLATAV RELKDFVSKN LTRAINKNKC DIDDLKMAV SFSQFNRRFL NVVRQFSENA GITPAISLDL MTDAELARAI SNMPTSAGQI KLMLENRAMV RRKGFGILIG V YGSSVIYM ...String: FVLGAIALGR ATAAAVTAGV AIAKTIRLES EVTAIKNALK TTNEAVSTLG NGVRVLATAV RELKDFVSKN LTRAINKNKC DIDDLKMAV SFSQFNRRFL NVVRQFSENA GITPAISLDL MTDAELARAI SNMPTSAGQI KLMLENRAMV RRKGFGILIG V YGSSVIYM VQLPIFGVID TPCWIVKAAP SCSEKKGNYA CLLREDQGWY CQNAGSTVYY PNEKDCETRG DHVFCDTAAG IN VAEQSKE CNINISTTNY PCKVSTGRNP ISMVALSPLG ALVACYKGVS CSIGSNRVGI IKQLNKGCSY ITNQDADTVT IDN TVYQLS KVEGEQHVIK GRPVSSSFDP IKFPPDQFNV ALDQVFENIE NSQAWVRKFD EILSSIEKG UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)