[English] 日本語
 Yorodumi
Yorodumi- EMDB-43144: MicroED structure of SARS-CoV-2 main protease (MPro/3CLPro) with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of SARS-CoV-2 main protease (MPro/3CLPro) with missing cone eliminated by suspended drop | ||||||||||||
 Map data Map data | 1.5 sigma | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cysteine Protease / Viral Protease / Viral Assembly / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / mRNA guanylyltransferase activity / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / snRNP Assembly / Translation of Replicase and Assembly of the Replication Transcription Complex / Replication of the SARS-CoV-2 genome / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell endosome / 3'-5'-RNA exonuclease activity / 5'-3' DNA helicase activity / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / symbiont-mediated suppression of host toll-like receptor signaling pathway / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / SARS-CoV-2 modulates host translation machinery / host cell Golgi apparatus / symbiont-mediated suppression of host NF-kappaB cascade / DNA helicase / symbiont-mediated perturbation of host ubiquitin-like protein modification / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / lyase activity / single-stranded RNA binding / viral protein processing / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / copper ion binding / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | electron crystallography / cryo EM / Resolution: 2.15 Å | ||||||||||||
 Authors Authors | Bu G / Gillman C / Danelius E / Hattne J / Nannenga BL / Gonen T | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2024 Journal: J Struct Biol X / Year: 2024Title: Eliminating the missing cone challenge through innovative approaches. Authors: Cody Gillman / Guanhong Bu / Emma Danelius / Johan Hattne / Brent L Nannenga / Tamir Gonen /  Abstract: Microcrystal electron diffraction (MicroED) has emerged as a powerful technique for unraveling molecular structures from microcrystals too small for X-ray diffraction. However, a significant hurdle ...Microcrystal electron diffraction (MicroED) has emerged as a powerful technique for unraveling molecular structures from microcrystals too small for X-ray diffraction. However, a significant hurdle arises with plate-like crystals that consistently orient themselves flat on the electron microscopy grid. If the normal of the plate correlates with the axes of the crystal lattice, the crystal orientations accessible for measurement are restricted because the crystal cannot be arbitrarily rotated. This limits the information that can be acquired, resulting in a missing cone of information. We recently introduced a novel crystallization strategy called suspended drop crystallization and proposed that crystals in a suspended drop could effectively address the challenge of preferred crystal orientation. Here we demonstrate the success of the suspended drop approach in eliminating the missing cone in two samples that crystallize as thin plates: bovine liver catalase and the SARS‑CoV‑2 main protease (Mpro). This innovative solution proves indispensable for crystals exhibiting systematic preferred orientations, unlocking new possibilities for structure determination by MicroED. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43144.map.gz emd_43144.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43144-v30.xml emd-43144-v30.xml emd-43144.xml emd-43144.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43144.png emd_43144.png | 126.1 KB | ||
| Filedesc metadata |  emd-43144.cif.gz emd-43144.cif.gz | 5.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43144 http://ftp.pdbj.org/pub/emdb/structures/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43144 | HTTPS FTP |
-Related structure data
| Related structure data |  8vd7MC  9bdjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43144.map.gz / Format: CCP4 / Size: 3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43144.map.gz / Format: CCP4 / Size: 3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1.5 sigma | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.713 Å / Y: 0.694 Å / Z: 0.709 Å | ||||||||||||||||||||||||||||||||||||
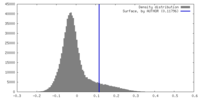
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 5 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 main protease (Mpro)
| Entire | Name: SARS-CoV-2 main protease (Mpro) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 main protease (Mpro)
| Supramolecule | Name: SARS-CoV-2 main protease (Mpro) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 3C-like proteinase nsp5
| Macromolecule | Name: 3C-like proteinase nsp5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: SARS coronavirus main proteinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.825547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGFRKMAFPS GKVEGCMVQV TCGTTTLNGL WLDDVVYCPR HVICTSEDML NPNYEDLLIR KSNHNFLVQA GNVQLRVIGH SMQNCVLKL KVDTANPKTP KYKFVRIQPG QTFSVLACYN GSPSGVYQCA MRPNFTIKGS FLNGSCGSVG FNIDYDCVSF C YMHHMELP ...String: SGFRKMAFPS GKVEGCMVQV TCGTTTLNGL WLDDVVYCPR HVICTSEDML NPNYEDLLIR KSNHNFLVQA GNVQLRVIGH SMQNCVLKL KVDTANPKTP KYKFVRIQPG QTFSVLACYN GSPSGVYQCA MRPNFTIKGS FLNGSCGSVG FNIDYDCVSF C YMHHMELP TGVHAGTDLE GNFYGPFVDR QTAQAAGTDT TITVNVLAWL YAAVINGDRW FLNRFTTTLN DFNLVAMKYN YE PLTQDHV DILGPLSAQT GIAVLDMCAS LKELLQNGMN GRTILGSALL EDEFTPFDVV RQCSGVTFQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #2: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 / Details: 0.1 M MES pH 6.5, 20% PEG 3350, 5% DMSO. |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 75.0 K / Max: 85.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number diffraction images: 420 / Average exposure time: 1.0 sec. / Average electron dose: 0.0025 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 0.0 µm / Nominal defocus min: 0.0 µm / Camera length: 2480 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN / Tilt angle: -40.0, 70.0 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.15 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystallography statistics | Number intensities measured: 123734 / Number structure factors: 14825 / Fourier space coverage: 95.5 / R merge: 25.6 / Overall phase residual: 0 / Phase error rejection criteria: none / High resolution: 2.15 Å Shell:
|
-Atomic model buiding 1
| Refinement | Protocol: OTHER / Target criteria: Rwork/Rfree gap of less than 5% |
|---|---|
| Output model |  PDB-8vd7: |
 Movie
Movie Controller
Controller












 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)