-Search query
-Search result
Showing 1 - 50 of 65 items for (author: hartman & ra)

EMDB-17104: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

EMDB-17105: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq4: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq5: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M
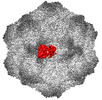
EMDB-26786: 
CPV Affinity Purified Polyclonal Fab A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
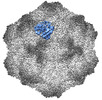
EMDB-26787: 
CPV Affinity Purified Polyclonal Fab B Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
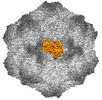
EMDB-26788: 
CPV Total-Fab Polyclonal A Site Fab
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
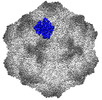
EMDB-26789: 
CPV Total-Fab Polyclonal B Site Fab (1 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ
EMDB-26790: 
CPV Total-Fab Polyclonal B Site Fab (2 of 2)
Method: single particle / : Hartmann SR, Hafenstein SL, Charnesky AJ

EMDB-26610: 
Cryo-EM structure of rabbit RyR1 in the presence of high Mg2+ and AMP-PCP in nanodisc
Method: single particle / : Nayak AR, Samso M
EMDB-15860: 
Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex
Method: single particle / : Pauwels E, Shewakramani NR, De Wijngaert B, Vermeire K, Das K

PDB-8b5l: 
Cryo-EM structure of ribosome-Sec61-TRAP (TRanslocon Associated Protein) translocon complex
Method: single particle / : Pauwels E, Shewakramani NR, De Wijngaert B, Vermeire K, Das K

EMDB-15863: 
Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147
Method: single particle / : Pauwels E, Shewakramani NR, De Wijngaert B, Vermeire K, Das K

PDB-8b6c: 
Cryo-EM structure of ribosome-Sec61 in complex with cyclotriazadisulfonamide derivative CK147
Method: single particle / : Pauwels E, Shewakramani NR, De Wijngaert B, Vermeire K, Das K

EMDB-14709: 
Cryo-EM structure of human NKCC1 (TM domain)
Method: single particle / : Nissen P, Fenton R, Neumann C, Lindtoft Rosenbaek L, Kock Flygaard R, Habeck M, Lykkegaard Karlsen J, Wang Y, Lindorff-Larsen K, Gad H, Hartmann R, Lyons J

PDB-7zgo: 
Cryo-EM structure of human NKCC1 (TM domain)
Method: single particle / : Nissen P, Fenton R, Neumann C, Lindtoft Rosenbaek L, Kock Flygaard R, Habeck M, Lykkegaard Karlsen J, Wang Y, Lindorff-Larsen K, Gad H, Hartmann R, Lyons J

EMDB-13416: 
Human voltage-gated potassium channel Kv3.1 (apo condition)
Method: single particle / : Chi G, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Savva C, Ragan TJ, Burgess-Brown NA, Duerr KL

EMDB-13417: 
Human voltage-gated potassium channel Kv3.1 (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Marsden B, MacLean EM, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

EMDB-13418: 
Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

EMDB-13419: 
Human voltage-gated potassium channel Kv3.1 (with EDTA)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Pike ACW, Marsden B, MacLean EM, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phh: 
Human voltage-gated potassium channel Kv3.1 (apo condition)
Method: single particle / : Chi G, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Savva C, Ragan TJ, Burgess-Brown NA, Duerr KL

PDB-7phi: 
Human voltage-gated potassium channel Kv3.1 (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Marsden B, MacLean EM, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phk: 
Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phl: 
Human voltage-gated potassium channel Kv3.1 (with EDTA)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Pike ACW, Marsden B, MacLean EM, Sader K, Burgess-Brown NA, Duerr KL

EMDB-12679: 
Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A
Method: single particle / : Qian P, Koblizek M

EMDB-12680: 
Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A
Method: single particle / : Qian P, Koblizek M

EMDB-12681: 
Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A
Method: single particle / : Qian P, Koblizek M

EMDB-12682: 
Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A
Method: single particle / : Qian P, Koblizek M

PDB-7o0u: 
Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A
Method: single particle / : Qian P, Koblizek M

PDB-7o0v: 
Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A
Method: single particle / : Qian P, Koblizek M

PDB-7o0w: 
Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A
Method: single particle / : Qian P, Koblizek M

PDB-7o0x: 
Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A
Method: single particle / : Qian P, Koblizek M

EMDB-24741: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

PDB-7ryj: 
Cryo EM analysis reveals inherent flexibility of authentic murine papillomavirus capsids
Method: single particle / : Hartmann SR, Hafenstein S

EMDB-22615: 
Cryo-EM structure of rabbit RyR1 in the presence of Mg2+ and AMP-PCP in nanodisc
Method: single particle / : Nayak AR, Samso M

EMDB-22596: 
Focused cryo-EM map of rabbit RyR1 central and transmembrane domains in the presence of Mg2+ and AMP-PCP in nanodisc
Method: single particle / : Nayak AR, Samso M

EMDB-11588: 
Structure of low-light grown Chlorella ohadii Photosystem I
Method: single particle / : Caspy I, Nelson N, Nechushtai R, Neumann E, Shkolnisky Y

EMDB-11589: 
Structure of high-light grown Chlorella ohadii photosystem I
Method: single particle / : Caspy I, Nelson N, Nechushtai R, Shkolnisky Y, Neumann E

EMDB-11640: 
Structure of small high-light grown Chlorella ohadii photosystem I
Method: single particle / : Caspy I, Nelson N

PDB-6zzx: 
Structure of low-light grown Chlorella ohadii Photosystem I
Method: single particle / : Caspy I, Nelson N, Nechushtai R, Neumann E, Shkolnisky Y

PDB-6zzy: 
Structure of high-light grown Chlorella ohadii photosystem I
Method: single particle / : Caspy I, Nelson N, Nechushtai R, Shkolnisky Y, Neumann E

PDB-7a4p: 
Structure of small high-light grown Chlorella ohadii photosystem I
Method: single particle / : Caspy I, Nelson N, Nechushtai R, Shkolnisky Y, Neumann E

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase
Method: subtomogram averaging / : Dietrich L

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria
Method: helical / : Parey K

EMDB-23081: 
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility
Method: single particle / : Hartmann SR, Goetschius DJ

PDB-7kzf: 
High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility
Method: single particle / : Hartmann SR, Goetschius DJ, Hafenstein S

EMDB-11692: 
Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP
Method: single particle / : Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet M, Castro-Hartman P, Qian P, Sader K, Dent K, Kimanius D, Sutherland JD, Lowe J, Barford D, Russo CJ

PDB-7aap: 
Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP
Method: single particle / : Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet M, Castro-Hartman P, Qian P, Sader K, Dent K, Kimanius D, Sutherland JD, Lowe J, Barford D, Russo CJ

EMDB-10223: 
Cryo-EM Structure of T. kodakarensis 70S ribosome
Method: single particle / : Matzov D, Sas-Chen A

EMDB-10224: 
Cryo-EM Structure of T. kodakarensis 70S ribosome in TkNat10 deleted strain
Method: single particle / : Matzov D, Sas-Chen A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
